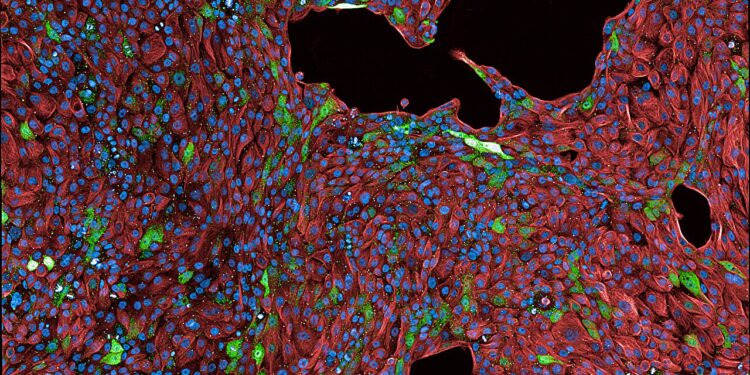
The LSD algorithm highlights lung cells that are actively responding (in green). Credit: Matthias Schmitt, Gargiulo Lab, Max Delbrück Center
All cells in our body have the same genetic code, yet they can differ in identity, functions and disease states. Distinguishing one cell from another in a simple way and in real time would prove invaluable to scientists trying to understand inflammation, infections or cancers.
Now, scientists at Germany’s Max Delbrück Center for Molecular Medicine have created an algorithm capable of designing such tools that reveal the identity and status of cells using segments of DNA called “locus control regions.” Synthetics” (sLCR). They can be used in various biological systems. The results, carried out by a team led by Dr. Gaetano Gargiulo, head of the molecular oncology laboratory, are reported in Natural communication.
“This algorithm allows us to create precise DNA tools to label and study cells, providing new insights into cellular behaviors,” says Gargiulo, lead author of the study. “We hope this research will open doors to a simpler, more scalable way to understand and manipulate cells.”
This effort began when Dr. Carlos Company, a former graduate student in the Gargiulo lab and co-first author of the study, began investing energy in automating the design of DNA tools and making them accessible to other scientists. He coded an algorithm to generate tools to help researchers understand fundamental cellular processes as well as disease processes such as cancers, inflammations and infections.
“This tool allows researchers to examine how cells transform from one type to another. It is particularly innovative because it compiles all the crucial instructions that direct these changes into a simple synthetic DNA sequence. In return “This simplifies the study of complex cellular behaviors in important areas like cancer research and human development,” says Company.
Algorithm to create a tailor-made DNA tool
The computer program is called “cis-regulatory synthetic DNA logic design” (LSD). Researchers enter known genes and transcription factors associated with the specific cellular states they want to study, and the program uses them to identify the DNA segments (promoters and enhancers) controlling activity in the cell of interest. This information is sufficient to discover functional sequences, and scientists do not need to know the precise genetic or molecular reason for a cell’s behavior; they just need to build the sLCR.
The program examines the human or mouse genome to find places where transcription factors are most likely to bind, says Yuliia Dramaretska, a graduate student in the Gargiulo lab and co-first author. It generates a list of 150 base pair long sequences that are relevant and likely act as active promoters and enhancers for the disease being studied.
“It’s obviously not a matter of giving a random list of these regions,” she says. “The algorithm classifies them and looks for the segments that will most effectively represent the phenotype you want to study.”
Like a lamp inside the cells
Scientists can then create a tool called a synthetic locus control region (sLCR), which includes the generated sequence followed by a DNA segment encoding a fluorescent protein.
“sLCRs are like an automated light that you can insert inside cells. This light only turns on under the conditions you want to study,” explains Dr. Michela Serresi, a researcher in the Gargiulo lab and co-first author . The color of the “lamp” can be varied to correspond to different states of interest, so scientists can look through a fluorescence microscope and immediately know the state of each cell by its color. “We can track with our eyes the color in a petri dish when we administer treatment,” says Serresi.
The scientists validated the utility of the computer program by using it to search for drugs in SARS-CoV-2-infected cells, as published last year in Scientists progress. They also used it to discover mechanisms involved in brain cancers called glioblastomas, for which no single treatment works.
“In order to find effective treatment combinations for specific cellular states in glioblastomas, you not only need to understand what defines these cellular states, but you also need to observe them as they arise,” says Dr. Matthias Jürgen Schmitt, researcher at the Gargiulo laboratory and co-first author, who used the laboratory’s tools to highlight their value.
Now imagine immune cells engineered in the lab as a gene therapy to kill a type of cancer. When infused into the patient, not all of these cells will function as intended. Some will be powerful while others might be in a dysfunctional state. The Gargiulo lab will use this system to study how these delicate cell-based cancer therapeutics behave during manufacturing.
“With the right collaborations, this method has potential to advance treatments in areas such as cancer, viral infections and immunotherapies,” says Gargiulo.
More information:
Carlos Company et al, Logical design of synthetic cis-regulatory DNA for genetic tracing of cellular identities and state changes, Natural communications (2024). DOI: 10.1038/s41467-024-45069-6
Provided by the Max Delbrück Center for Molecular Medicine
Quote: Using computer-generated DNA to study cellular identities (February 5, 2024) retrieved February 5, 2024 from
This document is subject to copyright. Except for fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.