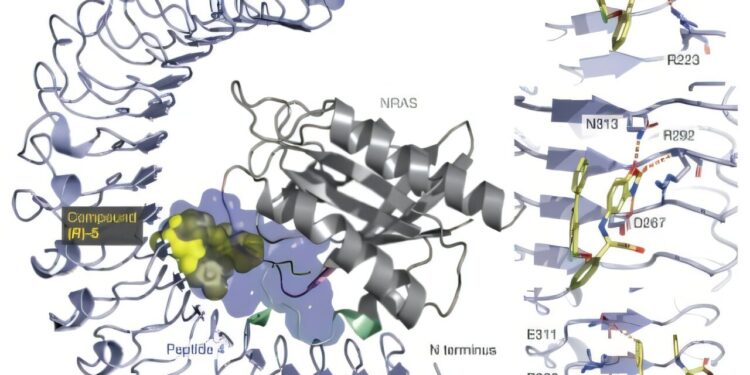
A) Representation of the SHOC2 linked to the Peptide 4 (Light Violet surface; PDB: 9BTN), compound (R) -5 (yellow spheres; PDB: 9BTP) and NRAS (Q61R) (Gray; PDB: 9BTM). b) The top, the compound (R) -5 interacts with R223 and Q269 via its carboxylic acid. Credit: Nature (2025). DOI: 10.1038 / S41586-025-08931-1
Researchers from Novartis Biomedical Research in the United States and Switzerland have identified a molecular target that could provide a new therapeutic path for cancers caused by NRAS mutations. The results suggest that interfere with specific protein interactions could disrupt the signaling pathways associated with uncontrolled cell proliferation in malignant tumors without targeted treatments.
Kras and Nras are members of the RAS gene family, which code for proteins that regulate cell growth and division by signaling pathways. The mutations of these genes disrupt normal signaling, leading to oncogenesis. Codon 12 mutations in KRAs are common in solid tumors, while codon 61 mutations in NRA are mainly associated with melanoma.
Targeted therapies have been developed for mutations in Kras (G12C) and Kras (G12D), but effective strategies for NRAS mutant cancers remain elusive. NRAS mutations represent 20 to 30% of melanoma cases, which represents a substantial therapeutic difference, in particular given the absence of approved targeted therapies for this subset.
With 50,000 new cases per year in the United States and Europe, NRAS mutant melanoma continues to present an unsecured clinical need. The identification of specific molecular dependencies associated with these mutations could reveal new therapeutic targets and clarify treatment strategies for malignant tumors focused on NRAS.
In the study, “targeting the SHOC2 – RAS interaction in rasant cancers”, published in NatureThe researchers led a CRISPR screen to the genome scale to identify genetic dependencies associated with NRAS mutations (Q61 *).
From BA / F3 cell lines designed to express KRAS and NRAS mutations to codons 12 and 61 were used to model the ONCOGENE RAS / MAPK dependence in the absence of other genetic alterations.
A Lenviral Arnm library on a genome scale has evaluated the growth dependencies of the Kras and Nras mutant lines under controlled conditions. The selective dependencies of mutants and isoforms were then confirmed using inhibitors of small molecule specific to Kras (G12C) and Kras (G12D). Subsequent analysis has focused on identifying unique genetic vulnerabilities with NRAS mutants (Q61 *).
SHOC2 has become a significant dependence in NRAS (Q61 *) – mutants. In the BA / F3 cellular lines, the Knockout Shoc2 mediated by the AGNA has led to a pronounced cellular lethality, with NRAS mutants (Q61 *) with increased sensitivity compared to the mutant lines of Kras (G12 *).
In the models of xenogreffe of melanoma, the exhaustion of SHOC2 led to an inhibition of tumor growth comparable to that observed after the Knockdown NRAS, suggesting that the targeting of SHOC2 could disrupt MAPK signaling in malignant tumors controlled by NRAS.
Analysis of the models of Mélanoma xenogreffe also demonstrated that the depletion of SHOC2 considerably reduced tumor growth in the mutant parameters of ANR (Q61 *). A reduced cell proliferation and a decrease in MAPK signaling were observed in vitro after the Knockdown SHOC2, aligning with the In Vivo results.
Mechanistic studies have confirmed that SHOC2 forms a direct interaction with the NRAS (Q61R) and KRAS (Q61R), with similar affinities. The structural optimization efforts have led to the identification of compounds of small molecules capable of linking Shoc2 and disturbing the SHOC2 – RAS interaction, with a compound demonstrating a dose -dependent inhibition of the RAS / MAPK signaling in the Mélanoma Mélanoma of ANR (Q61 *).
SHOC2 targeting in NRAS mutant cancer (Q61) presents a distinct therapeutic strategy of direct targeting of RAS, potentially preventing the signaling of cancer cells by SHOC2 – ARS interaction disturbances.
The identification of small SHOC2 molecule binders illustrates its drug potential in melanoma focused on NRAS, with wider implications for other RAS mutant cancers where SHOC2 regulates MAPK signaling.
More information:
Zachary J. Hausteman et al, targeting the SHOC2 – LAS interaction in the rasy cancers, Nature (2025). DOI: 10.1038 / S41586-025-08931-1
Hugo Lavoie et al, blocking a key node in the signaling of cancer unlocks the therapeutic potential, Nature (2025). DOI: 10.1038 / D41586-025-01136-6
© 2025 Science X Network
Quote: The CRISPR screen on a genome scale identifies a new target in melanoma focused on NRAS (2025, May 12) recovered on May 13, 2025 from
This document is subject to copyright. In addition to any fair program for private or research purposes, no part can be reproduced without written authorization. The content is provided only for information purposes.