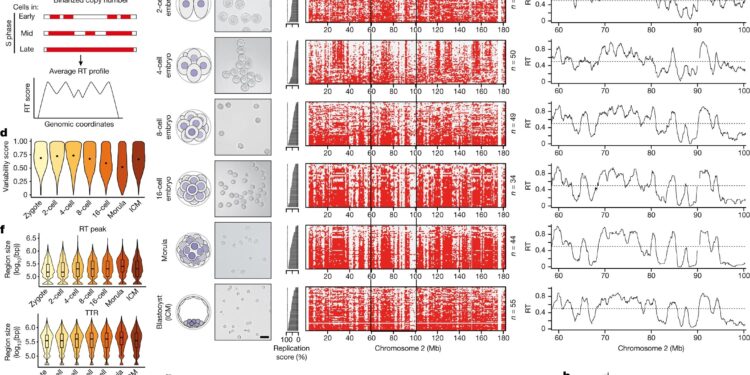
RT appears gradually during preimplantation development of the mouse. AOverview of single-cell Repli-seq used to generate RT profiles from single cells in mouse preimplantation embryos based on copy number variation. b, Embryo sampling scheme and corresponding images of dissociated blastomeres at each stage. The numbers of independent blastomere collections for each stage with similar results are as follows: zygote (3), 2 cells (4), 4 cells (3), 8 cells (3), 16 cells (3), morula. (2), ICM (4). Scale bar, 50 μm. vs, Heatmaps of single cells indicating replication status based on binarized copy numbers during preimplantation embryogenesis (red, replicated; gray, unreplicated). Cells are ranked based on their percentage of replicated genome (replication score), which indicates progression into S phase and is plotted as a bar graph on the left. d, Variability score during embryonic development; the score is 1 when 50% of the cells have replicated the genomic bin and 0 when all cells are either replicated (100%) or not replicated (0%). Each violin plot shows the distribution of scores for all genomic groups. eRT profiles of preimplantation embryos on a representative region of chromosome 2, denoted by a black rectangle in vs. The black line indicates RT profiles, calculated as the average of overlapping intervals defined by the genome-wide replication score. F,gSize (F) and the number (g) of replication exhibits RT peaks (also called initiation zones) and RT troughs (also called termination zones) during preimplantation development. The boxplots show the median and interquartile range (IQR), and the whiskers represent the lowest and highest values within a 1.5 × IQR interval. bp, base pair. h, Relative RT values centered on RT peaks during embryonic development relative to their neighboring regions. Note that the curves for the 2 and 4 cell stages overlap considerably and, to some extent, with those of the zygotes. Credit: Nature (2023). DOI: 10.1038/s41586-023-06872-1
The complex process of duplicating genetic information, called DNA replication, is central to the transmission of life from one cell to another and from one organism to another. This happens not only by copying genetic information; a well-orchestrated sequence of molecular events must also occur at the right time.
Scientists working with Professor Maria-Elena Torres-Padilla from Helmholtz Munich recently discovered a fascinating aspect of this process called “replication timing” (RT) and how special it is at the beginning of life. The new results are now published in Nature.
The process of DNA replication timing (RT) refers to the specific times when different regions of our genetic code are duplicated. Researchers at the Helmholtz Munich Institute for Epigenetics and Stem Cells implemented a technique called “Repli-seq” to delve deeper into the intimate relationship between RT and cell adaptability, cellular plasticity.
Intriguingly, they also discovered a new relationship between RT and how genes fold into three-dimensional structures inside the cell nucleus.
Starting with the earliest stage of an embryo, the zygote, the very beginning of an organism’s life, the researchers created a map of RT from this single-cell stage to the stage at which the embryo develops. implants in the mother’s uterus, called a blastocyst. The unexpected finding is that RT in the single-cell embryo is not very ordered, suggesting that genome duplications are very flexible in these early cells.
However, after the 4-cell stage, RT becomes more defined. A gradual process occurs, reflecting the gradual acquisition of changes in DNA and associated proteins, called chromatin marks, which indicate the activity and importance of genes in cellular functions.
Maria-Elena Torres-Padilla, corresponding author of the study, further explains: “This is remarkable, because it tells us that these early embryonic cells have a very ‘plastic’ genome duplication program. Because these first cells are totipotent, they can create every cell in our body. We believe that what we discovered in this study is one of the reasons why these cells are so remarkably capable of generating the whole body.
New discoveries about DNA replication may provide a tool for reprogramming cells. Dr. Tsunetoshi Nakatani, first author of the study, adds: “We can consider changing cell identity by changing its RT schedule to a more flexible schedule. »
The results further show that RNA polymerase, commonly known as the enzyme responsible for reading the genetic code and transcribing it into RNA, helps determine the exact program of RT, providing clues as to how such a program can be manipulated. in the future. .
The research team discovered that the three-dimensional structure of the genome takes shape first, and so the RT program is established. This is an exciting finding because it posits that the way our genome adapts to the three-dimensional space of the cell nucleus influences the flexibility of the RT program.
In conclusion, the timing of DNA replication is a fascinating piece of the puzzle of life’s grand narrative. This demonstrates how the precision of genetic replication is intimately linked to the ability of cells in the early embryo to generate other cell types in our body. As researchers continue to explore these connections, we gain a deeper understanding of the very essence of the transmission of life, from cell to cell, from organism to organism, and of what makes a cell capable of generate a new body.
More information:
Maria-Elena Torres-Padilla, Emergence of replication timing in early mammalian development, Nature (2023). DOI: 10.1038/s41586-023-06872-1
Provided by the Helmholtz Association of German Research Centers
Quote: Team discovers relationship between the timing of DNA replication and how genes fold into 3D structures inside the cell nucleus (December 20, 2023) retrieved December 21, 2023 from
This document is subject to copyright. Apart from fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.