Graphical summary. Credit: Nucleic acid research (2023). DOI: 10.1093/nar/gkad1164
Technological advances have allowed scientists to explore genetic control elements in depth, revealing the complexity of gene activation mechanisms in our genetic code. Emerging evidence challenges the simplistic view that cis-regulatory elements (CREs) are simply on/off switches for genes, highlighting their ability to exhibit complex behaviors, such as simultaneously enhancing gene activity and the initiation of gene transcription (e.g., a simultaneous enhancer and enhancer). promoter activities).
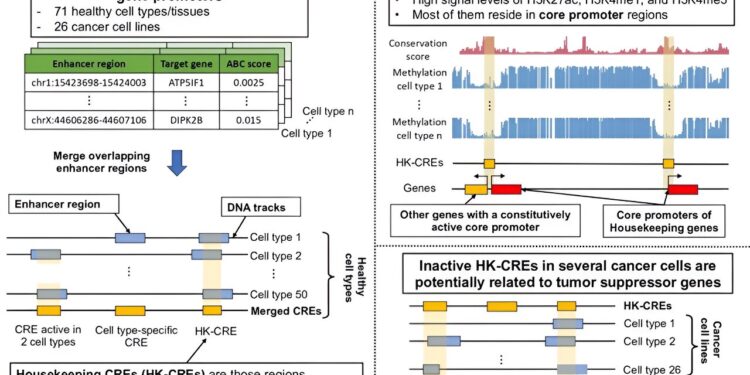
These switches are not only important for enhancing specific genes, but are also crucial for basic functions that keep our cells healthy. Now, a study in Japan has revealed the existence of approximately 11,000 vital genetic switches active in every cell type – housekeeping cis-regulatory elements (HK-CRE) – that play a role in maintaining of the stability and functioning of our cells, well beyond the regulation of housekeeping genes.
The research is published in Nucleic acid research. This study was carried out by scientists from the In Silico Functional Analysis Laboratory (Nakai-lab) at the Institute of Medical Sciences, University of Tokyo, Japan: Professor Kenta Nakai, head of the laboratory, and Dr. Martin Loza, assistant professor. , in collaboration with Dr Alexis Vandenbon, Associate Professor at the Institute of Life and Medical Sciences, Kyoto University, Japan.
Discussing his motivation behind this study, Dr. Loza says: “Given the significant association between cancer and mutations in epigenetic components, every little piece of information we obtain could be essential in the ongoing battle against this disease, which has tragically cost countless lives. Bioinformatic analyses, we aimed to highlight the profound impact of HK-CRE on fundamental cellular processes, including their potential as essential tumor suppressors.
The research team found that HK-CREs were not just limited to regulating well-studied housekeeping genes (HKGs), which made up less than 20% of the genes associated with these elements. Instead, these elements resided primarily in the core promoter regions of many more genes (approximately 8,000), indicating a broader regulatory role beyond the typical functions of housekeeping genes.
Using bioinformatics analyzes and leveraging various public datasets, the team validated the robustness of HK-CREs across 50 randomly selected healthy cell types, thereby confirming the location of HK-CREs in the genome. These elements were highly conserved, residing in unmethylated CpG-rich regions, a trait strongly associated with their housekeeping regulatory function.
Sharing his concerns about the analysis, Dr. Loza says, “By leveraging bioinformatics analyzes of multiomics data, we provide an approach to leverage publicly available datasets to explore diverse biological mechanisms. the time and financial investment required for in-depth studies involving new data.
The team noticed the complex cooperative interactions between housekeeping master promoters (HK-CPs), forming complex regulatory networks through promoter-promoter interactions. These observations suggest the significant influence of such interactions not only on HKGs but also on genes specific to different cell types.
Turning their attention to cancer cells, researchers discovered a subset of HK-CREs with reduced activity in various cancer subtypes due to aberrant methylation, particularly those related to zinc finger genes clustered in the subtelomeric regions of chromosome 19. By identifying genes such as ZNF135, ZNF154, ZNF667 and ZNF667-AS1, under the influence of these core promoters, the research suggests their potential as housekeeping tumor suppressor genes.
“The genes detected in our study showed reduced activity in several cancer cell lines, and survival analysis in various cancer projects revealed a significant increase in the probability of survival in various cancer types like pancreatic adenocarcinoma and uveal melanoma,” says Dr. Loza.
Essentially, the results of this research uncovered a previously unknown class of HK-CREs essential for cellular stability, extending their influence beyond the regulation of housekeeping genes.
“Our discovery of housekeeping tumor suppressor genes unveils a new avenue in cancer treatment, harnessing the intrinsic elements of each cell’s DNA. Future approaches to cancer treatment, focused on these housekeeping tumor suppressor genes, offer a unique solution that could potentially target a wide range of cancers, thereby avoiding the challenges associated with personalized medicine,” explains Dr. Loza.
“Our findings on cis-regulatory elements address a large gap in current knowledge regarding gene regulatory processes. We anticipate that our findings will improve the understanding of these processes and provide a valuable resource for researchers striving to discover elements inherent to the genome to fight against various diseases.
More information:
Martin Loza et al, Epigenetic characterization of core housekeeping promoters and their importance in tumor suppression, Nucleic acid research (2023). DOI: 10.1093/nar/gkad1164
Provided by the University of Tokyo
Quote: Study reveals crucial ‘housekeeping’ genetic elements and their potential in fighting cancer (January 5, 2024) retrieved January 5, 2024 from
This document is subject to copyright. Apart from fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.