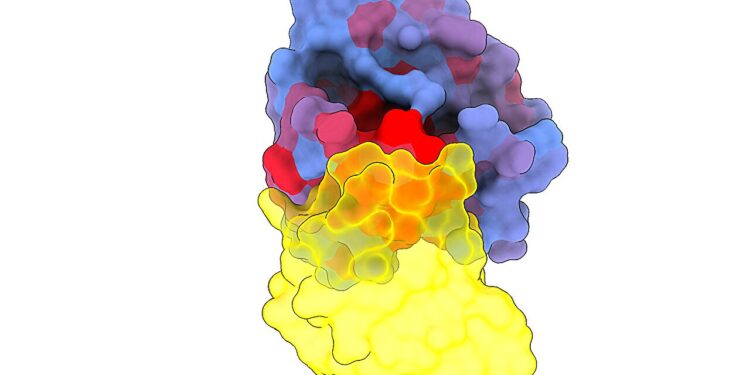
A three-dimensional image showing the human protein KRAS (blue) interacting with RAF1 (yellow), one of its main partners. The blue to red color gradient indicates an increasing potential for allosteric effects. Credit: Weng, Faure and Escobedo/Centro de Regulación Genómica
Researchers at the Center for Genomic Regulation in Barcelona, Spain, and the Wellcome Sanger Institute near Cambridge, United Kingdom, have comprehensively identified the allosteric control sites found in the KRAS protein. These are highly sought-after targets for drug development, representing secret vulnerabilities that can be exploited to control the effects of one of the most important causes of cancer. The study presents the first comprehensive control chart for any protein and is published in the journal Nature.
KRAS is one of the most frequently mutated genes in many types of cancers. It is found in one in ten human cancers, with a higher prevalence in fatal types such as pancreatic or lung cancers. It is called the “Death Star” protein because of its spherical shape and lack of a good site to target with drugs. For this reason, KRAS has historically been considered “non-drug” since its first discovery in 1982.
The only effective strategy to control KRAS has been to target its allosteric communication system. These are molecular signals that operate via a remote-controlled lock and key mechanism. To control a protein, you need a key (a chemical compound or drug) that can open a lock (active site). Proteins can also be influenced by a secondary lock (allosteric site) located elsewhere on their surface.
When a molecule binds to an allosteric site, it causes a change in the shape of the protein, which can alter the activity of the protein or its ability to bind other molecules, for example by changing the internal structure of its main lock.
Allosteric sites are often preferred for drug development because they provide greater specificity, thereby reducing the risk of side effects. They can also change a protein’s activity in more subtle ways, providing the opportunity to fine-tune its function. Drugs targeting allosteric sites are generally safer and more effective than drugs targeting active sites.
However, allosteric sites are very elusive. Despite four decades of research, tens of thousands of scientific publications, and more than three hundred published KRAS structures, only two drugs have been approved for clinical use: sotorasib and adagrasib. The drugs work by attaching to a pocket adjacent to the active site, inducing an allosteric conformational change in the protein that prevents it from being activated.
“It took decades to produce an effective drug against KRAS, in part because we lacked tools to identify allosteric sites on a large scale, meaning we were searching for therapeutic target sites in the dark. In this study “, we demonstrate a new approach that can systematically map allosteric sites for entire proteins. For the purposes of drug discovery, it’s like turning on the lights and laying bare the many ways we can control a protein,” explains Dr. André Faure, scientist at the Center for Genomic Regulation and co-author of the study.
Four promising targets for safer, more effective drugs
The study authors mapped the allosteric sites using a technique called deep mutational analysis. This involved creating more than 26,000 variations of the KRAS protein, changing just one or two building blocks (amino acids) at a time. The team checked how these different variations of KRAS bind to six other proteins, including those essential for KRAS to trigger cancer. The researchers used AI software to analyze the data, detect allostery, and identify the location of known and novel therapeutic target sites.
“The unique selling point of our method is its scalability. In this work alone, we performed more than 22,000 biophysical measurements, a number similar to the total ever achieved for all proteins before we began exploiting the remarkable advances in DNA sequencing and synthesis methodologies. is a huge acceleration and demonstrates the power and potential of the approach,” says Chenchun Weng, first author of the study and postdoctoral researcher at the Center for Genomic Regulation.
The technique revealed that KRAS has many more strong allosteric sites than expected. Mutations in these sites inhibited the protein’s binding to its three major partners, suggesting that broad inhibition of KRAS activity is possible. A subset of these sites is particularly interesting because they are located in four different easily accessible pockets on the protein surface and represent promising targets for future drugs.
The authors of the study highlight one in particular, “pocket 3,” as being particularly interesting. This pocket is located far from the active site of KRAS and has therefore received very little attention from pharmaceutical companies.
The researchers also found that small changes to KRAS can dramatically change its behavior with its partners, making the proteins preferred over each other. This has important implications because it could lead to new strategies for controlling aberrant KRAS activity without hindering its normal function in non-cancerous tissues.
Sparing normal versions of KRAS means fewer side effects and safer, more effective treatments. Researchers could also use this knowledge to delve deeper into the biology of KRAS and explain how the protein behaves in various scenarios, which could be key to determining its role in different types of cancer.
A new plan to drug the “untamed”
The study provides the first-ever comprehensive map of allosteric sites for any complete protein in any species. Research shows that with the right tools and techniques, such as those used to map KRAS, new vulnerabilities can be discovered for many medically important proteins that have historically been considered “undruggable.”
“The big challenge in medicine is not knowing which proteins cause diseases, but how to control them. Our study represents a new strategy for targeting these proteins and accelerating the development of drugs to control their activity. The nature of targeting protein allosteric sites means that the resulting drugs will likely be safer and more effective treatments than we currently have,” concludes ICREA Research Professor Dr. Ben Lehner, lead author of the Center study. for Genomic Regulator and the Wellcome Sanger Institute.
More information:
The energetic and allosteric landscape for KRAS inhibition, Nature (2023). DOI: 10.1038/s41586-023-06954-0
Provided by the Center for Genomic Regulation
Quote: Secret Vulnerabilities of Cancer’s ‘Death Star’ Protein Revealed (December 18, 2023) Retrieved December 18, 2023 from
This document is subject to copyright. Except for fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.