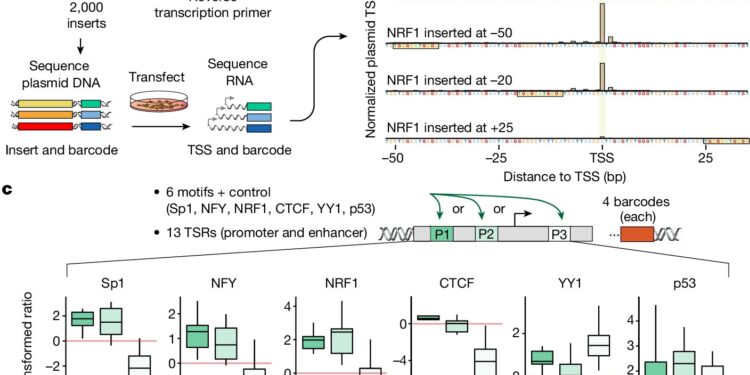
TSS capture of thousands of TSR variants confirms position-dependent function of TFs. Credit: Nature (2024). DOI: 10.1038/s41586-024-07662-z
A newly discovered code in DNA, called a “spatial grammar,” holds the key to understanding how gene activity is encoded in the human genome.
This revolutionary discovery, identified by researchers from Washington State University and the University of California, San Diego and published in Naturehas revealed a long-postulated hidden spatial grammar embedded in DNA. This research could reshape scientists’ understanding of gene regulation and how genetic variation can influence gene expression in development or disease.
Transcription factors, the proteins that control when genes in the genome are turned on or off, play a crucial role in this code. Long thought to be activators or repressors of gene activity, this research shows that the function of transcription factors is much more complex.
“Contrary to what you find in textbooks, transcription factors that act as true activators or repressors are surprisingly rare,” said Sascha Duttke, a WSU assistant professor who led much of the research at WSU’s School of Molecular Biosciences in the College of Veterinary Medicine.
Instead, scientists have discovered that most activators can also function as repressors.
“If you remove an activator, your assumption is that you lose activation,” said Bayley McDonald, a WSU graduate student who was part of the research team. “But that was only true 50 to 60 percent of the time, so we knew something was wrong.”
Looking more closely, the researchers found that the function of many transcription factors was highly position-dependent.
They found that the spacing between transcription factors and their position relative to a gene’s transcription start point determined the level of gene activity. For example, transcription factors can activate gene expression when positioned upstream or before a gene’s transcription start point, but inhibit its activity when positioned downstream or after a gene’s transcription start site.
“It’s the spacing, or ‘ambience,’ that determines whether a given transcription factor acts as an activator or a repressor,” Duttke said. “It just goes to show that, like learning a new language, to learn how gene expression patterns are encoded in our genome, we need to understand both its words and its grammar.”
“By incorporating this newly discovered ‘spatial grammar,'” Christopher Benner, an associate professor at UC San Diego, predicts that scientists will be able to gain a deeper understanding of how mutations or genetic variations can affect gene expression and contribute to disease.
“The potential applications are vast,” Benner said. “At a minimum, this will change the way scientists study gene expression.”
More information:
Sascha H. Duttke et al., Position-dependent function of human sequence-specific transcription factors, Nature (2024). DOI: 10.1038/s41586-024-07662-z
Provided by Washington State University
Quote:Scientists Discover New Code Governing Gene Activity (2024, August 20) retrieved August 20, 2024 from
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without written permission. The content is provided for informational purposes only.