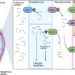
The illustration shows a human protein and amino acid code in the background. The new FRET X technology is able to identify proteins using protein fingerprints. The Chirlmin Joo Lab obtains these unique fingerprints by finding part of the complete amino acid code (the C and K highlighted among the blue letters). Credit: Delft University of Technology
In a study published in Nature Nanotechnology, scientists from Delft University of Technology present a new technique for identifying proteins. Proteins perform essential functions in our cells, while also playing a crucial role in diseases like cancer and COVID-19 infection. Researchers identify proteins by reading the fingerprint and comparing the fingerprint to patterns in a database.
With this new technology, researchers can identify individual, intact and complete proteins, preserving all their information. This can shed light on the mechanisms behind many different diseases and enable earlier diagnosis.
Incomplete IKEA project
“The study of proteins in cells has been a hot topic for decades and has led to enormous progress, allowing researchers to have a much better idea of the type of proteins present and the function they perform,” explains Mike Filius, lead researcher. author of the paper.
Currently, scientists use a method called mass spectrometry to identify proteins. The most common mass spectrometry approach is the “bottom-up” approach, in which full-length proteins are broken into smaller fragments, called peptides, which are then measured by the mass spectrometer. Based on the data from these small fragments, a computer reconstructs the protein.
Filius says: “It’s a bit like your typical IKEA project, where you always end up with spare parts that you don’t really know how to fit them into. But in the case of protein, these spare parts may actually contain very little. valuable information, for example on whether or not such a protein has a harmful disease-causing structure.
Protein fingerprinting
“To identify a protein, you don’t need to know all the amino acids, the building blocks of a protein. Instead, you try to get enough information to be able to identify the protein using a base of data as a reference, similar to how the police can discover the identity of a suspect through their fingerprints,” explains Filius.
“In previous work, we showed that each protein has a unique fingerprint, just like the human analogue. We realized that we only need to know the location of a few of the amino acids in a protein to generate a unique fingerprint from the fingerprint. which we can identify the protein,” adds Raman van Wee, a Ph.D. candidate who participated in the research.
Finding Protein in a Haystack
“We can detect these amino acids through molecules that light up under a microscope and are attached to small pieces of DNA that bind very specifically to a certain amino acid,” says Van Wee. This way, the team can determine the location of the amino acid very quickly with high precision.
“Because the sensitivity of this new technique, called FRET a tiny amount of sample,” Filius said. said. This is important because it allows patient samples to be measured in the event of illness.
“In our paper we show that we can detect small amounts of proteins characteristic of Parkinson’s disease or COVID-19 infection,” says Filius.
“Although other approaches are being explored to identify proteins, ours focuses on identifying intact, individual proteins in a complex mixture. We can look for a needle in a haystack,” adds Van Wee.
Towards early diagnosis of the disease
Although promising, the research still requires substantial development, which the Chirlmin Joo Lab is eager to work on. The research group spoke with several stakeholders from clinical laboratories and the biopharmaceutical industry and learned that they are truly excited about the revolutionary potential of this technology.
They are also working to launch a start-up to develop FRET X into a highly sensitive protein detection platform. This platform can diagnose diseases in the early stages, improving the effectiveness of potential treatments.
“This revolutionary technique deciphers the protein code and opens up exciting possibilities for early detection of diseases,” explains Chirlmin Joo, project supervisor.
More information:
Mike Filius et al, Complete fingerprinting of a single protein molecule, Nature Nanotechnology (2024). DOI: 10.1038/s41565-023-01598-7
Provided by Delft University of Technology
Quote: Scientists develop new technology to identify individual full-length human proteins (February 15, 2024) retrieved February 15, 2024 from
This document is subject to copyright. Apart from fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.