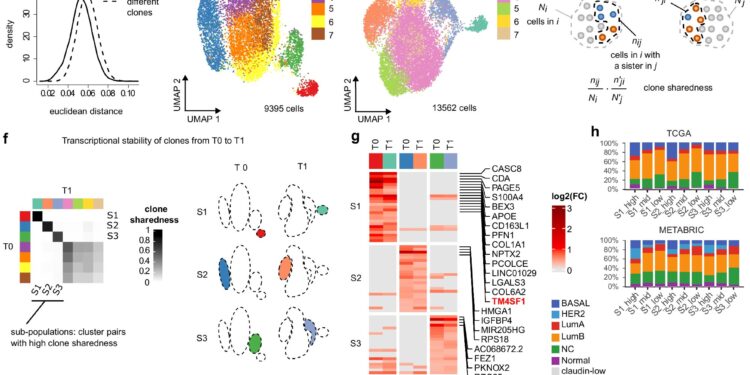
Lineage tracing identifies transcriptionally stable TNBC cell subpopulations. Credit: Natural communications (2024). DOI: 10.1038/s41467-024-51424-4
Using a genetic barcoding system and a new single-cell sequencing method, a research team from the Istituto Italiano di Tecnologia (IIT-Italian Institute of Technology) in Milan has developed an approach to identify the cells responsible for triggering tumors and metastases, particularly in breast cancer.
Using these same techniques, researchers also discovered which of these cells are capable of resisting chemotherapy, even before these characteristics appear in patients. The results were published in the journal Natural communications.
Barcodes are typically used to identify commercial products and track their movements. However, the IIT research group at the Milan Center for Genomic Sciences, led by Dr. Francesco Nicassio, used them for a less conventional purpose: to label cancer cells and track their evolution over time.
The study specifically focused on triple negative breast cancer, which accounts for approximately 20% of breast cancer cases. This form of cancer is difficult to treat, representing a significant challenge for both therapy and research.
Genetic tags have been assigned to individual cancer cells from triple-negative breast cancer, allowing researchers to trace their evolutionary path during tumor development and growth. This approach allowed the creation of a distinct and recognizable profile for selected cells in the resulting cancer.
“Identifying so-called tumor-initiating cells is not easy, but thanks to close multidisciplinary collaboration and the use of cutting-edge multi-omics technologies, including single-cell sequencing, we were able to achieve this result,” explains Dr. . Nicassio, who led the research.
“Based on the molecular features we identified, we could select which tumor cells are capable of forming metastases and which are capable of developing drug resistance.”
The next step was to study the genetic aspects of the monitored cells, including their genetic, epigenetic and transcriptional characteristics. The researchers developed a multi-omics method, an innovative approach to study these characteristics simultaneously.
The results revealed that epigenetic features – modifications that, without altering the DNA or RNA sequence, can influence gene expression – play a critical role in both initial tumor development and formation of metastases.
“We identified a ‘pro-metastatic epigenome’, a kind of molecular signature present in the primary tumor that marks the most aggressive cells,” adds Dr Matteo Marzi from the Center for Genomic Sciences at IIT Milan, the one of the authors of the study. the paper.
Using these molecular signatures, the researchers were able to classify the cells as more or less aggressive and distinguish them from another population of cells that develop drug resistance due to genetic mutations.
“Our work was primarily about finely characterizing the molecular profiles of individual cells, using innovative technologies to observe and understand what we could previously only assume,” explains co-author Dr Francesca Nadalin, a researcher at IIT of Milan and at the European Institute of Bioinformatics. (EMBL-EBI) in Cambridge, United Kingdom.
“The results suggest that specific regions of the genome may be involved in the development of specific properties of cancer, such as tumor proliferation or resistance to chemotherapy.”
The research team aims to investigate further, with the aim of eventually introducing these findings into clinical practice. These results could serve as a cornerstone for new early diagnosis methods and innovative therapeutic treatments.
The next steps are to validate the results on a wider range of cultured cells and to better understand the link between molecular profiles and the underlying causes of metastasis and drug resistance.
More information:
F. Nadalin et al, Multi-omics lineage tracing predicts transcriptional, epigenetic and genetic determinants of cancer evolution, Natural communications (2024). DOI: 10.1038/s41467-024-51424-4
Provided by the Italian Institute of Technology
Quote: Researchers show that tumor evolution is written in the genome (October 15, 2024) retrieved October 15, 2024 from
This document is subject to copyright. Except for fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for informational purposes only.