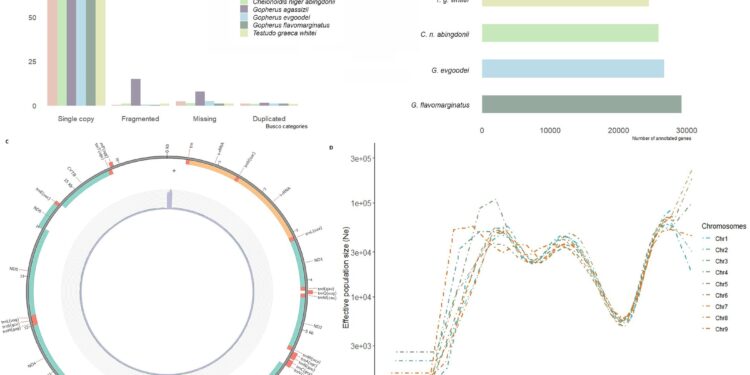
Representation of the main results of the genome assembly. Credit: PLOS ONE (2024). DOI: 10.1371/journal.pone.0303408
Like many endangered species of tortoises, the wandering tortoise did not have a complete genome. For the first time, researchers from the Departments of Ecology at the Miguel Hernández University of Elche (UMH) and the University of Alicante (UA) have managed to sequence the genome of the wandering tortoise, using as a reference the genome of another closely related native American tortoise.
The results, published in PLOS ONEwill enable the scientific community to support the conservation of these endangered animals.
The graeca tortoise (Testudo graeca) is one of the most emblematic species of land turtles in the Mediterranean basin. In the Iberian Peninsula, graeca tortoises have two main populations: one in the southeast, from the north of Almería to the south of Murcia, and the other in the Doñana National Park. The species is endangered in Andalusia and is included in the catalogue of endangered species of the Regional Ministry of Murcia and the Ministry of the Environment.
“Understanding the genetic diversity of animals can be very useful for the conservation of species like the pointed-thighed tortoise, because the more we know, the better we can understand how these animals have adapted to their environment or what their capacity is to cope with climate change,” explains Andrea Mira Jover, researcher at UMH and lead author of the study.
In recent years, Mira-Jover adds, conservation biology has used a promising tool: genome sequencing. A genome is the complete set of DNA instructions contained in a cell. Sequencing a genome involves reading all the genetic information representative of a species, identifying specific genes, for example, and organizing them into chromosomes.
The description of the genome of this species constitutes a crucial scientific step, because very few turtles have been described at this level.
“These results will be a starting point to better understand the evolutionary history of the species and resolve questions related to its life cycle, such as the secret of its longevity,” emphasizes UA researcher Roberto Rodríguez-Caro. In addition, he adds, the publication of this reference genome will provide key tools for its global conservation, since the species is classified as vulnerable by the International Union for Conservation of Nature (IUCN) and requires concrete measures to preserve its populations in the future.
Rodríguez-Caro, from the UA Department of Ecology, has been studying this species for 15 years, collecting information on its ecology, conservation and genetics in collaboration with various national and international research centers.
Different techniques exist for obtaining complete genomes, depending on whether the genetic information is read in long or short fragments.
“If all DNA were a novel, some techniques could read long sentences, while others would identify individual words,” Mira-Jover says. Long-read techniques are more efficient at assembling genomes de novo (organizing DNA sequences without using a prior reference), but they are still too expensive. However, other methods can obtain complete genomes using short-read techniques by referencing the genomes of closely related species.
“In this case, the novel is written using individual words instead of long sentences,” says the UMH researcher.
This method, called “reference assembly,” is particularly useful in species that evolve slowly, that is, have a low rate of genetic change. They maintain the same order of genes, called “highly syntenic groups.” Using this new metaphor, if only individual words are available to write the genetics of a species, sentences from a similar book can be consulted to complete the genome.
“Turtles, scientifically known as chelonians or testudines, are an example of slow-evolving organisms,” explains Eva Graciá, a researcher at UMH, head of the study and president of the Spanish Association of Herpetology. “Chelonians are an ancient and diverse taxonomic group, which includes freshwater, marine and land turtles, but their genomic organization is very similar,” she points out, adding that “turtles have evolved very slowly throughout history, and their genes are similar and located in the same place on the chromosomes.”
Tortoises (Testudinidae) are the most threatened family. However, only five reference genomes are available compared to 33 for marine and freshwater turtles. Faced with this situation, the scientific community needs more resources to contribute to the conservation of tortoise populations.
For this reason, researchers from the UMH Department of Ecology have generated the first reference genome at the chromosomal level of the goose-footed tortoise, using short-read sequencing techniques. They used the known genome of Gopherus evgoodei, the Sinaloa spiny tortoise, native to the deserts of the United States and Mexico.
If we imagine the DNA double helix as a spiral staircase, each step of the staircase would be made up of what are called “base pairs” containing smaller molecules. The size of a complete genome is measured by the number of base pairs. For example, the human genome has 3.2 billion base pairs containing about 25,000 genes.
Using various bioinformatics techniques, the researchers analyzed a 2.2 billion base pair genome of the goose-footed tortoise containing nearly 26,000 genes. They also performed a demographic reconstruction to understand the evolutionary history of the species.
“This analysis suggests an evolutionary model very similar to what we have already observed with other techniques in previous studies,” emphasizes UMH ecology professor Andrés Giménez, author of the publication.
This reference genome will help answer questions about the evolutionary history of the spurred tortoise and allow genes of interest to be studied in future studies. It will also be a valuable tool for making better conservation decisions. In addition, the rest of the scientific community will have access to the spurred tortoise genome, making a significant contribution to a field with limited resources.
More information:
Andrea Mira-Jover et al., Leveraging reference-guided assembly in a slowly evolving lineage: application to Testudo graeca, PLOS ONE (2024). DOI: 10.1371/journal.pone.0303408
Provided by Miguel Hernandez University of Elche
Quote:Researchers sequence the genome of the spur-thighed tortoise (2024, September 10) retrieved September 10, 2024, from
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without written permission. The content is provided for informational purposes only.