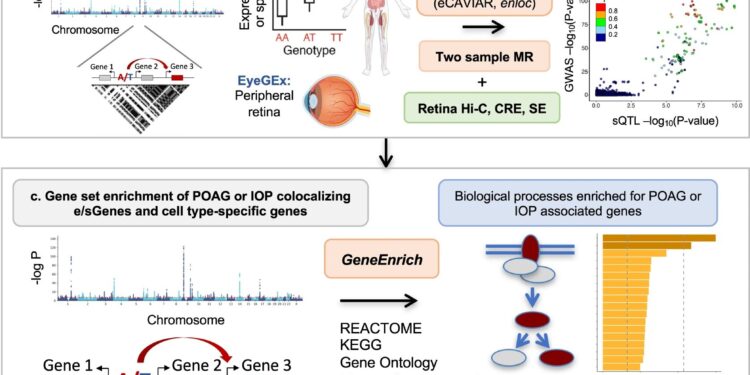
Analysis workflow from POAG and IOP GWAS to causal regulatory mechanisms, genes, pathways and cell types. A Genome-wide POAG and IOP associations (known and modest associations) were tested for enrichment of expression and splicing quantitative trait loci (e/sQTL) in GTEx tissues and retina compared to permuted null sets of variants matched on confounding factors, using QTLEnrich (unilateral). In cases where enrichment was found, the lower bound number of e/sQTLs in a given tissue likely to be true trait associations was estimated using a true positive rate derived from empirically ((π1)({\pi }_{1} ) approach. b Putative causal genes were prioritized according to the known genome-wide association study (GWAS) locus of POAG and IOP by applying two colocalization methods (eCAVIAR, enloc) to all e/sQTLs from 49 GTEx tissues and retinal eQTLs that overlapped each locus, followed by two-sample Mendelian randomization (MR). Overlap of colocalizing GWAS loci and e/sQTL with Hi-C (3D chromosome conformation capture), cisRegulatory element (CRE) and super-enhancer (SE) regions of the human retina were used to further prioritize causal genes. Human and eye images were created with BioRender.com. vs All target genes for significant colocalization of e/sQTLs (e/sGenes) or cell type-specific genes by trait were tested for enrichment in signaling and metabolic pathways (Reactome, KEGG), gene ontologies and mouse phenotype ontologies using GeneEnrich (unilateral). The Manhattan plot of the POAG cross-ancestry GWAS meta-analysis was generated using QMplot ( d Significantly colocalized e/sGenes were tested for enrichment in specific cell types in glaucoma-relevant ocular tissue mononuclear RNA sequence data, using ECLIPSER (one-tailed). Cell type specific genes were defined with cell type fold change >1.3 and FDR <0.1 per tissue. The significance of cell type specificity per GWAS locus defined for a given trait was assessed against a null distribution of loci associated with unrelated, non-ocular traits using a Bayesian Fisher's exact test. Genes mapped to GWAS loci with a cell type specificity score greater than 95thA percentile of null locus scores was proposed to contribute to the trait in the enriched cell type. e Cell type enrichment for POAG and IOP GWAS was corroborated using two regression-based methods that assess the cell type specificity of trait associations considering all associations at scale. of the genome: stratified score-LD (S-LDSC) and MAGMA regression. Credit: Natural communications (2024). DOI: 10.1038/s41467-023-44380-y
Although primary open-angle glaucoma (POAG) is the leading cause of blindness in people over the age of 55, there is still no cure for the disease and its biological mechanisms are not well understood. Elevated intraocular pressure (IOP) is a major risk factor for the disease, but many glaucoma patients have normal eye pressure and still lose vision.
In a new study published last month in Natural communicationsMass Eye and Ear researchers, led by Ayellet Segrè, Ph.D., conducted a comprehensive study combining genetic findings from a large genome-wide association meta-analysis of cross-ancestry, led by Janey Wiggs, MD, PhD and a broad meta-analysis of IOP with gene regulation studies and single-cell expression measurements in glaucoma-relevant ocular tissues.
In doing so, they discovered key genes, biological processes, and cell types that may affect POAG pathogenesis in an IOP-independent and IOP-dependent manner.
Using integrative analyses, they identified hundreds of genes and regulatory effects underlying more than 100 loci associated with POAG and/or IOP that may contribute to glaucoma risk due to high levels of glaucoma. modified gene expression. These genes are enriched in biological pathways involved in disease mechanisms, including elastic fiber formation and extracellular matrix organization, vascular development, and neuronal processes.
Additionally, using single-nucleus gene expression data from the aqueous humor outflow tract, retina, optic nerve head, and surrounding posterior tissues, as part of a A whole-eye cell atlas led by Josh Sanes, Ph.D. of the Department of Molecular and Cellular Biology at Harvard University, researchers identified known and less well-established cell types in which genetic dysregulation can affect the degeneration of the optic nerve.
This included fibroblasts in the conventional and unconventional outflow tracts, astrocytes in the retina and optic nerve head, oligodendrocytes and vascular cells in the optic nerve head, and fibroblasts in the surrounding region of the optic nerve head. head of the optic nerve.
The pathways identified in their analysis may impact various structures of the eye, pointing to many potential mechanisms that may interact with different genes and cell types that may contribute to POAG. These results provide new insights into gene expression and post-transcriptional gene regulation that could improve glaucoma drug design.
“Our work has generated new insights into the mechanisms of POAG, which could inform the development of new therapies targeting IOP reduction and neuroprotection,” Segrè said.
“For example, this research suggests that targeting supporting neuronal cells, in addition to retinal ganglion cells, may be important to consider in the design of new drugs and cellular therapies. Through our ongoing work aimed at detect genetic regulation of gene expression in glaucoma-relevant ocular tissues, hopefully in the future to provide a more comprehensive understanding of POAG risk and IOP variation.
More information:
Andrew R. Hamel et al, Integrating gene regulation and single-cell expression with GWAS prioritizes glaucoma causative genes and cell types, Natural communications (2024). DOI: 10.1038/s41467-023-44380-y
Provided by Mass. General Brigham
Quote: Researchers identify genes and cell types that may play a causal role in the formation of primary open-angle glaucoma (February 16, 2024) retrieved February 16, 2024 from
This document is subject to copyright. Apart from fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.