Credit: Cell Reports Medicine (2024). DOI: 10.1016/j.xcrm.2023.101359
Doctors have nearly a dozen new targeted drugs to treat patients with acute myeloid leukemia, or AML, but three out of four patients still die within five years. Some patients succumb in just a month or two, despite the battery of medications used to treat this aggressive blood disease, in which blood cells fail to grow properly.
A new study draws on a field of science known as proteogenomics to try to improve the outlook. In an article published on January 16 in Cell Reports MedicineScientists report new findings about how drug resistance develops in some AML patients and how doctors might one day stop or slow the process.
The research comes from a team of researchers at the Department of Energy’s Pacific Northwest National Laboratory and Oregon Health and Science University. For nearly a decade, OHSU and PNNL researchers have worked together to fill a critical gap in our knowledge about the onset of cancer and other diseases. At one end of the spectrum, our body’s genes can malfunction, creating mutations that can be harmful or even fatal. On the other end of the spectrum is a real person whose life is affected, or even ended, as a result.
What happens between genes and a person’s health?
The answer: a dizzying number of complex molecular processes that scientists are struggling to understand. At the center are the body’s proteins and a field of study called proteogenomics.
Sort data with machine learning
The PNNL-OHSU team is studying thousands of proteins that may play a role in AML. Proteins are the body’s molecular workhorses, transporting nutrients and other supplies between cells, turning genes on or off, and maintaining dozens of basic bodily processes.
Although genes get the glory, they don’t do much directly to keep our bodies functioning. That’s the job of proteins. For nearly 20 years, study author Karin Rodland of OHSU, formerly of PNNL, has been a pioneer in exploring the role of proteins in health and disease, building a program with colleagues from OHSU and PNNL to study AML.
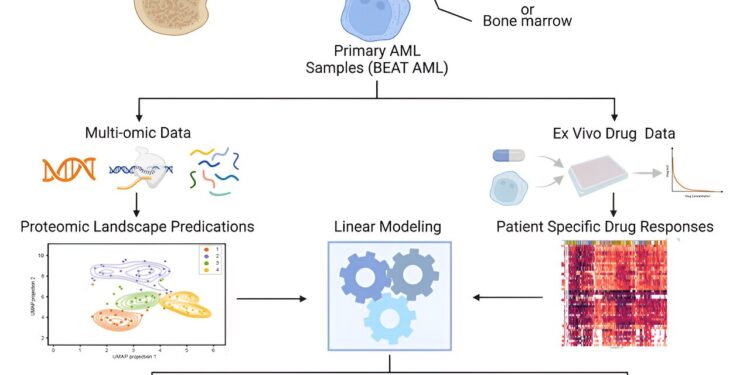
In the latest study, a team led by Sara Gosline, a data scientist and computational biologist at PNNL, performed a comprehensive study of protein activity in 210 AML patients. In total, the team measured the levels of nearly half a million protein pieces from more than 9,000 proteins in the patients’ blood samples.
The team combined these results with extensive data already known about the disease: the genes and mutations involved, the molecular messengers that indicate which genes are active and the effects of 46 drugs on AML patients, as well as information on disease progression in these patients. the patients.
“We were able to examine patterns of drug responses in hundreds of people by including protein and gene measurements, which gave us a level of detail that had not been possible in previous studies,” said Gosline. “This is a great example of how we are able to apply our growing knowledge of protein signaling models and machine learning to benefit patients in the future.”
Gosline and his colleagues, including first author James Pino of PNNL, deployed artificial intelligence using multiple machine learning algorithms to make sense of the data.
Overcoming drug resistance
Although the study provided a wealth of data about what happens in the body of an AML patient, one finding stood out, indicating a possible way to avoid or delay drug resistance in some patients.
The team showed that quizartinib treatment, approved last year to treat AML, can change the way cancer cells respond to other drugs often used in combination to treat patients.
Specifically, the team found that when patients on quizartinib stop responding to venetoclax, doctors might consider switching to another drug, panobinostat. It’s an example of how proteogenomic information could change the road map that doctors use to determine which drugs patients receive at different stages of the disease.
“The difficulty is that the cancer continues to evolve,” Gosline said. “You hit the tumor with a drug and the tumor changes. This is what happens when patients develop drug resistance and the drugs stop working. Our study helps us understand exactly how this happens and what causes it. could be done in response. Which medicine is best for whom?”
AML poses a particular challenge, said study author Cristina Tognon of OHSU.
“When you treat a tumor with a drug, you put pressure on the tumor cells as they try to find a way to escape that pressure and survive. This is a big problem in AML patients. What is even more difficult is that in AML there are many mutations at work; the disease does not have just one flavor,” said Tognon, associate research professor and scientific director of the laboratory. Drunk at OHSU.
Ultimately, the team focused on 147 specific proteins and molecular locations called phosphosites that play a key role in determining which proteins are turned on and off.
Using only the protein data, the team sorted the samples into four distinct groups that predicted patient outcomes. Patients whose samples placed them in one of the groups had a better prognosis than the others, surviving well over five years. Doctors hope this type of information will eventually become available in the clinic. This would allow some patients who do not need aggressive therapies that cause serious side effects to avoid them while ensuring that patients with the worst prognosis are treated as aggressively as possible.
“There is potential for clinical applications derived from this work, for example diagnostics, such as protein biomarkers to predict responses to therapies, and the design of new drug combinations that could outperform current ones,” said Jeff Tyner from OHSU, professor of medicine at the OHSU School of Medicine and the Knight Cancer Institute.
This work is the latest in more than 200 studies examining protein activity in many forms of cancer, including colon, brain, endometrial, brain, blood and ovarian cancers. An OHSU-PNNL team discussed the emerging role of proteins in treating patients with precision medicine in a recent article in the Annual pharmacology and toxicology exams. Increasingly, scientists are using proteomics – the study of proteins – to bridge the gap between genomics (the study of genes) and phenotype (phenotypes or observable characteristics).
More information:
James C. Pino et al, Mapping the proteogenomic landscape helps predict drug response in acute myeloid leukemia, Cell Reports Medicine (2024). DOI: 10.1016/j.xcrm.2023.101359
Sunil K. Joshi et al, Mass spectrometry-based proteogenomics: new therapeutic opportunities for precision medicine, Annual Review of Pharmacology and Toxicology (2023). DOI: 10.1146/annurev-pharmtox-022723-113921
Provided by Pacific Northwest National Laboratory
Quote: Proteins suggest a pathway to reduce drug resistance in a form of cancer (February 2, 2024) retrieved February 2, 2024 from
This document is subject to copyright. Except for fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.