Credit: Cell Reports Medicine (2023). DOI: 10.1016/j.xcrm.2023.101227
Before a drug can be used to treat a disease, it must go through a lengthy and expensive testing process to prove both its safety and effectiveness. By repurposing previously approved drugs, researchers can sometimes reduce the time and expense of the first step. Repurposed drugs fail as often as new drugs in clinical trials designed to study their effectiveness, according to Penn State researchers.
To improve the success rate of drug reuse and determine effective treatment doses, Penn State researchers developed a model that predicts effective doses for reused drugs. Their results were published in Cell Reports Medicine.
“The nice thing about the process of repurposing a drug is that there is no need to go back and double-check that it is safe for a patient,” said the corresponding author Justin R. Pritchard, associate professor of biomedical engineering at Penn State. “But when we actually looked at the numbers, it turned out that just because you had to skip that first step and reduce a lot of the costs associated with drug development, the ultimate success rate of these reuse efforts was about as good as trying to start from scratch with a new drug.”
Paper co-author and Penn State biomedical engineering doctoral student Scott M. Leighow said the low success rate was largely due to a problem understanding the drug’s dose during preliminary testing.
“A researcher might think, ‘This drug works really well for leukemia. Maybe it will be a good drug to treat pancreatic cancer or diabetes or COVID,'” he said. “What often ends up happening is they take this drug and throw enough of it at the cells in a plate until they see the desired effect, then call it a potential therapy, even though the quantity may not be reasonable to give to a patient.
The other problem, Leighow said, is that isolated cells in a dish may behave differently from cells surrounded by proteins and tissues in a patient’s body.
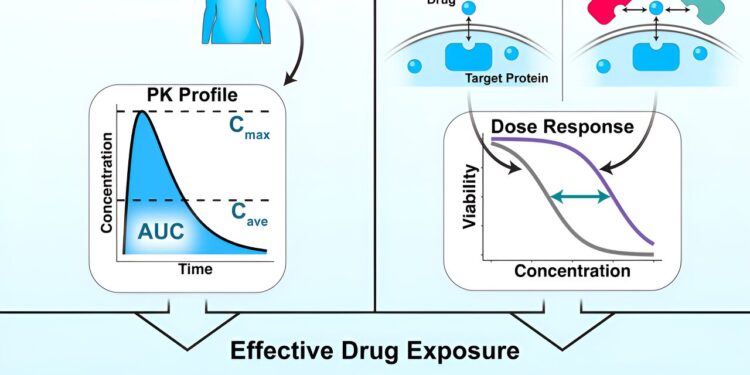
To address this problem, researchers developed a computer model using existing data on how a leukemia drug actually performed in real patients. They then fed the model information with their own experimental data on drug-disease interactions for different concentrations on isolated cells in a dish. Specifically, the researchers tested the drug on isolated cancer cells and on cancer cells in a solution of sticky proteins, which exist in the blood and keep some of the drug away from its target.
By dividing the first number – the percentage of the drug that reached the target cancer cells in tests done with sticky proteins – by the second, taken from tests done without sticky proteins, they obtain the serum displacement factor. They could then multiply that number by the drug concentration found in a patient’s blood sample to produce a corrected concentration.
“It turns out that it’s a very good indicator of how effective this drug is in this disease setting,” Leighow said. “We call this effective drug exposure in a patient, and it’s what allows us to translate the concentration values between a patient and the cells in a dish.”
Leighow said they could ask the model for the concentration corrected for a particular drug-disease association, which serves as a threshold.
“Experimental performance above that threshold that we would call clinically effective, and performance below that, we would say that the drug does not work in that setting,” Leighow said. “We found that we could take the same number that the model had given us and try to evaluate data that the model had never seen before, and ask, ‘How well does this same threshold predict whether Does the drug work in this setting?” It turned out that this same number worked very well in completely different diseases.”
For this research, the team used leukemia data, but when they applied it to lung cancer and gastrointestinal stromal tumors, they saw similar success rates (90% accuracy) and discovered that it could use similar thresholds to predict drug effectiveness.
“Even though that threshold leaves a little bit of disease-specific wiggle room, for the computer to be able to tell us precisely what that number was, it needed that broader biological context,” Leihow said. “We had to give him the right numbers, whereas before that wasn’t the case.”
Although this research focused on finding effective doses for drug repurposing, the researchers said they believe this model could be used to design new drugs in a similar class in the future.
More information:
Chuan Liu et al, Excessive concentrations of kinase inhibitors in translational studies hinder effective drug repurposing, Cell Reports Medicine (2023). DOI: 10.1016/j.xcrm.2023.101227
Provided by Pennsylvania State University
Quote: Predicting correct dosage can improve drug reuse success (January 12, 2024) retrieved January 12, 2024 from
This document is subject to copyright. Except for fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.