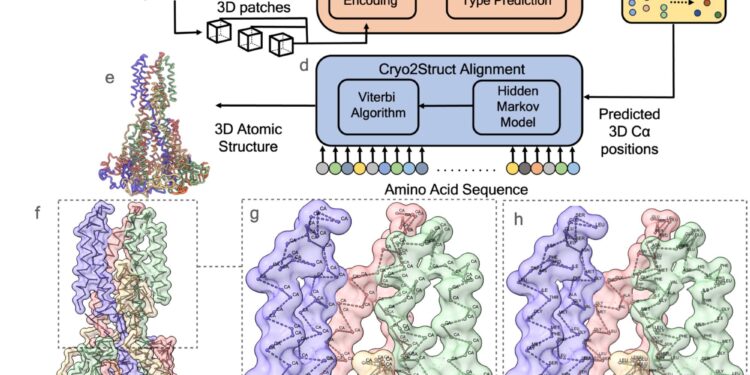
A look at Cryo2Struct’s automated prediction workflow. Credit: Nature Communications (2024). DOI: 10.1038/s41467-024-49647-6
A University of Missouri researcher has created a computer program that can unlock the mysteries of how proteins work together, giving scientists valuable insights to better prevent, diagnose and treat cancer and other diseases.
Jianlin “Jack” Cheng of Mizzou’s College of Engineering and his student, Nabin Giri, have developed a tool called Cryo2Struct that uses artificial intelligence (AI) to construct the three-dimensional atomic structure of large protein complexes, work recently published in Nature CommunicationsThe model uses data from images of frozen molecules captured by powerful microscopes or from cryo-electron microscopy (cryo-EM) images.
“Cryo-EM is now a revolutionary and essential technology for determining large protein structures and assemblies in cells,” said Cheng, professor emeritus of electrical engineering and computer science.
“But building protein structures from Cryo-EM data requires a lot of labor and human intervention, making it time-consuming and difficult to reproduce. Our technique is fully automated and generates more accurate structures than existing methods.”
Predicting proteins
To understand the importance of this work, you need to know a little about proteins and the decades-long struggle to understand them.
Proteins are the building blocks of life. They are made up of chains of amino acids that fold together to form three-dimensional shapes. These shapes determine how a protein functions.
For more than 50 years, this folding process has baffled researchers.
Cheng was one of the first to apply deep learning, a type of AI, to this problem. In 2012, he presented an AI-based model that proved that deep learning could predict protein structures. This work paved the way for groundbreaking advances, including Google’s AlphaFold, now considered the world’s most accurate tool for predicting protein structures.
But predicting the structure of a single protein is only half the battle. In the real world, proteins work together like molecular machines that perform complex biological functions. Understanding the interactions between proteins is critical because they determine how diseases develop and help scientists determine the best way to treat them.
Decrypt the code
Cheng’s Cryo2Struct works a bit like a detective solving a case without any clues.
The system analyzes cryo-EM images and identifies individual atoms and their positions within a protein complex, even without prior knowledge of the structure. The system can then assemble these atoms into a complete 3D model of protein complexes, providing insights into how proteins function.
“Our technology allows scientists to determine and build a structure from cryo-EM data,” Cheng said. “Once you have that structure and understand its functions, you can design drugs to counteract any defective function of a protein complex to make it work properly.”
In a related article published in Communication ChemistryCheng and his student, Alex Morehead, explored another AI method called diffusion modeling, which models how molecular structures evolve from random noise to well-defined shapes. These methods can help scientists generate and optimize small molecules, including drugs, and determine how and where those drugs bind to proteins.
“For example, I have a drug and I want it to be more effective for certain patients,” Cheng says. “Now I can use AI to modify and optimize it.”
Mizzou’s interdisciplinary resources helped make this breakthrough possible. Cheng is a researcher at NextGen Precision Health, where he has access to cryo-EM and high-resolution electron microscopy.
“The opportunities Mizzou provides to collaborate with other researchers and utilize cutting-edge equipment are unprecedented,” he said. “At NextGen, we are all working to advance highly individualized health care, and technologies like Cryo2Struct will help make that possible.”
More information:
Nabin Giri et al, De novo modeling of protein atomic structure for cryoEM density maps using 3D transformer and HMM, Nature Communications (2024). DOI: 10.1038/s41467-024-49647-6
Alex Morehead et al, Full geometric diffusion for 3D molecule generation and optimization, Communication Chemistry (2024). DOI: 10.1038/s42004-024-01233-z
Provided by the University of Missouri
Quote: Paving the way for new treatments with a tool that builds 3D structures of protein complexes (2024, September 23) retrieved September 24, 2024 from
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without written permission. The content is provided for informational purposes only.