Credit: Natural aging (2024). DOI: 10.1038/s43587-023-00548-1
Autophagy, which declines with age, may hold more mysteries than researchers previously suspected. In the January 4 issue of Natural agingIt was noted that scientists from the Buck Institute, Sanford Burnham Prebys and Rutgers University have discovered possible new functions for various autophagy genes, which could control different forms of elimination, including misfolded proteins , and ultimately affect aging.
“While this is very basic research, this work reminds us that it is critical for us to understand whether we have the full history of different genes linked to aging or age-related diseases ” said Professor Malene Hansen, Ph.D., Buck’s scientific director, who is also the co-senior author of the study.
“If the mechanism we discovered is conserved in other organisms, we believe it could play a more important role in aging than previously estimated and could provide a method for improving lifespan.”
These new observations provide another perspective on what was traditionally thought to happen during autophagy.
Autophagy is a cellular “housekeeping” process that promotes health by recycling or removing damaged DNA and RNA and other cellular components in a multi-step degradation process. It has been shown to play a key role in the prevention of aging and age-related diseases, including cancer, cardiovascular disease, diabetes and neurodegeneration. In particular, research has shown that autophagy genes are responsible for prolonged lifespan in various long-lived organisms.
The classic explanation of how autophagy works is that cellular “waste” to be processed is sequestered in a vesicle surrounded by a membrane, and ultimately delivered to lysosomes for degradation. However, Hansen, who has studied the role of autophagy in aging for most of her career, was intrigued by accumulating evidence indicating that this was not the only process in which autophagy genes can work.
“There has been this increasingly widespread idea over the last few years that genes present in the early stages of autophagy are ‘moonlighting’ in processes outside of this classic lysosomal degradation,” he said. -she declared. Furthermore, although it is known that multiple autophagy genes are required to increase lifespan, the tissue-specific roles of specific autophagy genes are not well defined.
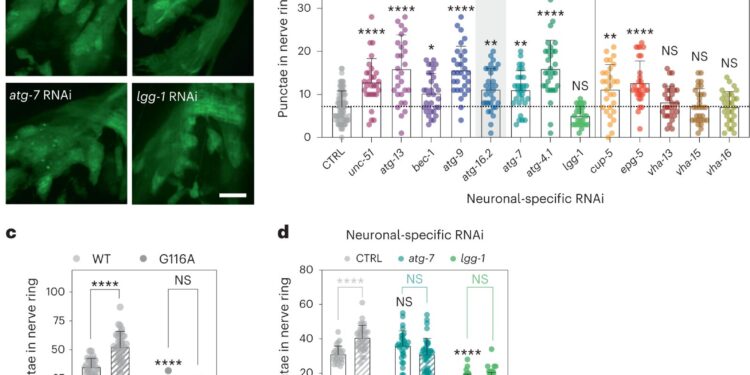
To further study the role that autophagy genes play in neurons – a key cell type for neurodegenerative diseases – the team analyzed Caenorhabditis elegans, a small worm frequently used to model the genetics of aging and which has a very well studied nervous system.
The researchers specifically inhibited the functioning of autophagy genes at each step of the process in the animals’ neurons and found that neuronal inhibition of early-acting, but not late-acting, autophagy genes prolonged the duration of life. These initial observations were made in Dr. Hansen’s laboratory at Sanford Burnham Prebys in La Jolla, California, before joining the Buck Institute in 2021.
An unexpected aspect was that this extension of lifespan was accompanied by a reduction in aggregated proteins in neurons (an increase is associated with Huntington’s disease, for example) and an increase in the formation of what we call them exophes. These giant vesicles extruded from neurons were identified in 2017 by Dr. Monica Driscoll, collaborator and professor at Rutgers University.
“Exophers are thought to be essentially another cellular method of waste disposal, a mega-bag of waste,” said Dr. Caroline Kumsta, co-senior author and assistant professor at SBP. “When there is too much waste accumulating in neurons, or when the normal ‘internal’ waste disposal system is broken, cellular waste is then dumped into these exophes.”
Interestingly, worms that formed exophes had reduced protein aggregation and lived significantly longer. This finding suggests a link between this mass elimination process and overall health, Kumsta said. The team discovered that this process depends on a protein called ATG-16.2.
The study identified several new functions of the autophagy protein ATG-16.2, including in exophe formation and lifespan determination, leading the team to speculate that this protein plays a non-traditional and unexpected role in the aging process. If this same mechanism works in other organisms, it could provide a method for manipulating autophagy genes to improve neuronal health and increase lifespan.
“But first we need to learn more, including how ATG-16.2 is regulated and whether this is relevant in a broader sense, in other tissues and other species,” Hansen said. The Hansen and Kumsta teams plan to track a number of longevity models, including nematodes, mammalian cell cultures, human blood and mice.
“Knowing whether there are multiple functions around autophagy genes like ATG-16.2 will be extremely important for developing potential therapies,” Kumsta said. “This is very basic biology right now, but this is where we are in knowing what these genes do.”
The traditional explanation that aging and autophagy are linked due to lysosomal degradation may need to be expanded to include additional pathways, which should be targeted differently to treat diseases and associated problems. “It will be important to know either way,” Hansen said. “The implications of such additional functions could lead to a potential paradigm shift.”
More information:
The autophagy protein ATG-16.2 and its WD40 domain mediate the beneficial effects of inhibiting early-acting autophagy genes in C. elegans’ neurons, Natural aging (2024). DOI: 10.1038/s43587-023-00548-1 www.nature.com/articles/s43587-023-00548-1
Provided by the Buck Institute for Research on Aging
Quote: New roles of autophagy genes in cellular waste management and aging (January 4, 2024) retrieved on January 4, 2024 from
This document is subject to copyright. Apart from fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.