R2R. Credit: Natural biotechnology (2024). DOI: 10.1038/s41587-023-02082-2
Sequencing all of a cell’s RNA can reveal a lot of information about how that cell works and what it’s doing at any given time. However, the sequencing process destroys the cell, making it difficult to study ongoing changes in gene expression.
An alternative approach developed at MIT could allow researchers to track these changes over extended periods of time. The new method, based on a non-invasive imaging technique known as Raman spectroscopy, does not damage cells and can be performed repeatedly.
Using this technique, the researchers showed that they could monitor embryonic stem cells as they differentiated into several other cell types over several days. This technique could enable the study of long-term cellular processes such as cancer progression or embryonic development, and could one day be used for the diagnosis of cancer and other diseases.
“With Raman imaging, you can measure many more points in time, which can be important for studying cancer biology, developmental biology, and a number of degenerative diseases,” says Peter So, professor of engineering. biology and mechanics at MIT, director of MIT Laser. Biomedical Research Center and one of the authors of the article.
Koseki Kobayashi-Kirschvink, a postdoctoral fellow at MIT and the Broad Institute of Harvard and MIT, is the lead author of the study, which appears in Natural biotechnology. The paper’s lead authors are Tommaso Biancalani, a former Broad Institute scientist; Jian Shu, assistant professor at Harvard Medical School and associate member of the Broad Institute; and Aviv Regev, executive vice president of Genentech Research and Early Development, who is on leave from his faculty position at the Broad Institute and the Department of Biology at MIT.
Gene expression imaging
Raman spectroscopy is a non-invasive technique that reveals the chemical composition of tissues or cells by shining near-infrared or visible light onto them. The MIT Biomedical Laser Research Center has been working on biomedical Raman spectroscopy since 1985, and recently, So and other members of the center have developed techniques based on Raman spectroscopy that could be used to diagnose breast cancer or measure the blood sugar.
However, Raman spectroscopy alone is not sensitive enough to detect signals as small as changes in the levels of individual RNA molecules. To measure RNA levels, scientists typically use a technique called single-cell RNA sequencing, which can reveal which genes are active in different cell types in a tissue sample.
In this project, the MIT team sought to combine the advantages of single-cell RNA sequencing and Raman spectroscopy by training a computational model to translate Raman signals into RNA expression states.
“RNA sequencing gives you extremely detailed information, but it is destructive. Raman is non-invasive, but it doesn’t tell you anything about RNA. So the idea of this project was to use machine learning to combine the strength of both modalities, allowing you to understand the dynamics of gene expression profiles at the single-cell level over time,” explains Kobayashi-Kirschvink.
To generate data to train their model, the researchers treated mouse fibroblast cells, a type of skin cell, with factors that reprogram the cells to become pluripotent stem cells. During this process, cells can also transition to several other cell types, including neural and epithelial cells.
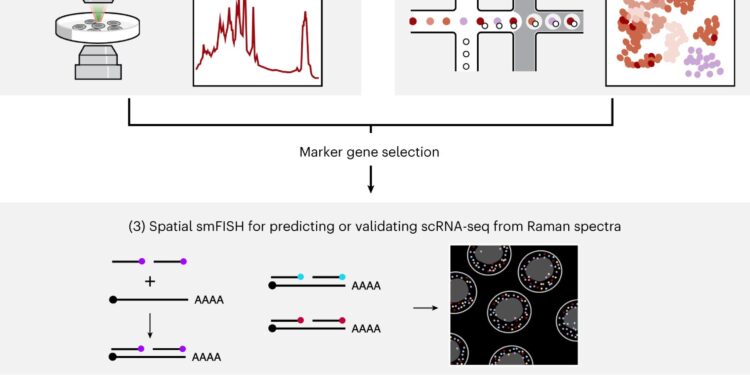
Using Raman spectroscopy, the researchers photographed the cells at 36 time points over 18 days as they differentiated. After each image was taken, the researchers analyzed each cell using single-molecule fluorescence in situ hybridization (smFISH), which can be used to visualize specific RNA molecules in a cell. In this case, they looked for RNA molecules encoding nine different genes whose expression patterns vary between cell types.
These smFISH data can then serve as a link between Raman imaging data and single-cell RNA sequencing data. To establish this link, the researchers first trained a deep learning model to predict the expression of these nine genes based on the Raman images obtained from these cells.
Next, they used a computer program called Tangram, previously developed at the Broad Institute, to link the smFISH gene expression patterns to the whole genomic profiles they had obtained by performing single-cell RNA sequencing on cells from the sample.
The researchers then combined these two computer models into one they call Raman2RNA, which can predict the entire genomic profiles of individual cells based on Raman images of the cells.
Monitoring cell differentiation
The researchers tested their Raman2RNA algorithm by tracking mouse embryonic stem cells as they differentiated into different cell types. They took Raman images of the cells four times a day for three days and used their computer model to predict the corresponding RNA expression profiles of each cell, which they confirmed by comparing them to the sequencing measurements. ‘RNA.
Using this approach, researchers were able to observe the transitions that occurred in individual cells as they differentiated from embryonic stem cells into more mature cell types. They also showed that they could track the genomic changes that occur when mouse fibroblasts are reprogrammed into induced pluripotent stem cells, over a two-week period.
“This is a demonstration that optical imaging provides additional information that makes it possible to directly follow the lineage of cells and the evolution of their transcription,” explains So.
The researchers now plan to use this technique to study other types of cell populations that change over time, such as aging cells and cancer cells. They are currently working with cells grown in a laboratory dish, but in the future they hope this approach can be developed as a potential diagnostic for use in patients.
“One of the biggest advantages of Raman is that it is a label-free method. It is still a long way off, but there is potential for human translation, which could not be achieved using invasive techniques to measure genomic profiles,” says Jeon. Woong Kang, researcher at MIT and also author of the study.
More information:
Prediction of single-cell RNA expression profiles in living cells by Raman microscopy with Raman2RNA, Natural biotechnology (2024). DOI: 10.1038/s41587-023-02082-2. On bioRxiv: DOI: 10.1101/2021.11.30.470655
Provided by the Massachusetts Institute of Technology
This story is republished courtesy of MIT News (web.mit.edu/newsoffice/), a popular site that covers news about MIT research, innovation and education.
Quote: Non-invasive technique reveals how cells’ gene expression changes over time (January 10, 2024) retrieved January 10, 2024 from
This document is subject to copyright. Apart from fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.