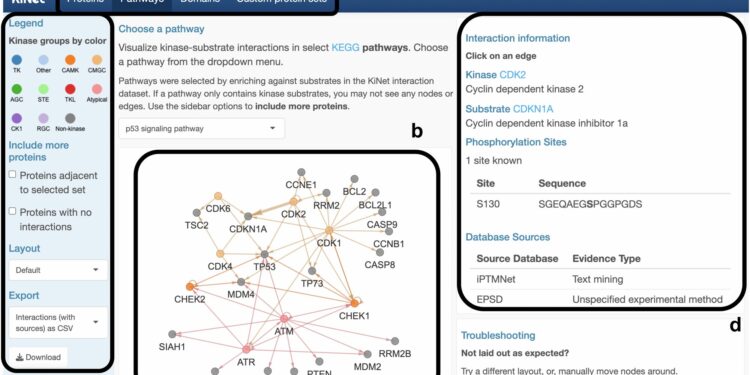
KiNet features. Credit: Biology and applications of npj systems (2024). DOI: 10.1038/s41540-024-00442-5
Researchers from the Icahn School of Medicine at Mount Sinai introduced KiNet, an interactive web portal designed to explore kinase-substrate interactions in human cellular systems. These interactions play a critical role in modulating complex signaling pathways that control various cellular processes, such as cell growth, differentiation, and response to environmental stimuli.
By integrating data from multiple public databases, KiNet allows the scientific community to visualize and study these interactions in systemic contexts, thereby improving understanding of kinase functions and their implications in diseases such as cancer, neurodegenerative disorders and cardiovascular diseases.
KiNet details are published in Biology and applications of npj systems.
Kinases are enzymes that modify other proteins by adding phosphate groups, a process called phosphorylation. This modification regulates protein activity, localization, and function, ultimately influencing various cellular activities. Disruption of these kinase-substrate interactions is often linked to diseases, making them major targets for therapeutic intervention.
Despite the abundance of data on kinases and their substrates, much of this information is scattered across multiple databases, making interpretation difficult in the context of cellular systems. KiNet solves this problem by aggregating data from major databases and providing a user-friendly platform for researchers to explore these interactions comprehensively.
“Kases and their substrates play critical roles in cellular signaling pathways, and the ability to visualize these interactions in system-level contexts opens new avenues for understanding their roles in health and disease,” said Gaurav Pandey, Ph.D., professor of genetics and genomic sciences at Icahn Mount Sinai and co-senior author of the paper. “KiNet not only aggregates existing data, but also provides an interactive platform that will facilitate drug discovery and systems-level analysis. »
KiNet’s ability to visualize kinase-substrate interactions within pathways, domain families, and custom protein sets provides researchers with a powerful tool for knowledge discovery. By focusing on systemic contexts, such as the network of interactions in signaling pathways, users can gain a more comprehensive view of how these important proteins function. This capability holds promise for a wide range of applications, including drug discovery, where understanding the complex relationships between kinases and substrates can reveal potential targets for therapeutic intervention.
In addition to its use in basic research, KiNet is poised to contribute to the development of new therapies. Many current drugs target kinases because of their central role in diseases such as cancer, where abnormal kinase activity drives tumor growth. KiNet’s interactive approach to visualizing kinase interactions can help researchers identify new drug targets and understand the effects of potential treatments on complex signaling pathways.
“KiNet represents a unique resource for the scientific community, bringing together diverse data sets and providing an accessible platform for visualizing kinase-substrate interactions,” said Avner Schlessinger, Ph.D., professor of pharmacological sciences at Icahn Mount Sinai and co-senior author. paper. “We anticipate that KiNet will be a valuable tool for advancing both basic research and clinical applications, particularly in drug discovery and personalized medicine.”
More information:
John AP Sekar et al, A web portal for exploring kinase-substrate interactions, Biology and applications of npj systems (2024). DOI: 10.1038/s41540-024-00442-5
Provided by Mount Sinai Hospital
Quote: New web portal enables drug discovery and systems-level analysis of critical kinase-substrate interactions (October 3, 2024) retrieved October 3, 2024 from
This document is subject to copyright. Except for fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for informational purposes only.