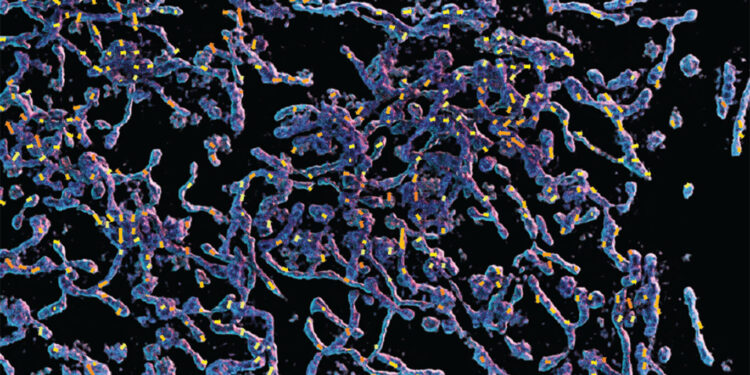
3D rendering of mitochondria overlaid on the motion map. The 3D volume was captured by a SIM 3D microscope in the Biological Imaging Facility and reconstructed by NSTM using the Advanced Bioimaging Center compute cluster. Credit: Ruiming Cao
Imaging microscopic samples requires capturing multiple sequential measurements and then using computer algorithms to reconstruct a single, high-resolution image. This process can work well when the sample is static, but if it moves, as is often the case with living biological specimens, the final image may be blurry or distorted.
Now, Berkeley researchers have developed a method to improve the temporal resolution of these dynamic samples. In a study published in Natural methodsThey present a new computational imaging tool, called Neural Space-Time Model (NSTM), which uses a small, lightweight neural network to reduce motion artifacts and resolve motion trajectories.
“The challenge of imaging dynamic samples is that the reconstruction algorithm assumes a static scene,” said lead author Ruiming Cao, a Ph.D. student in bioengineering.
“NSTM extends these computational methods to dynamic scenes by modeling and reconstructing motion at every moment. This reduces artifacts caused by motion dynamics and allows us to see these ultra-rapid changes within a sample.”
According to the researchers, NSTM can be integrated into existing systems without requiring expensive additional hardware. And it’s very effective. “NSTM has been shown to provide an order of magnitude improvement in temporal resolution,” Cao said.
The open source tool also allows the reconstruction process to operate on a finer time scale. For example, the computational process of imaging reconstruction may involve capturing approximately 10 or 20 images to reconstruct a single super-resolved image.
But using neural networks, NSTM can model how the object changes over those 10 or 20 frames, allowing scientists to reconstruct a super-resolved image on the time scale of one frame rather than every 10 or 20 images.
“Basically, we use a neural network to model the dynamics of the sample over time, so we can reconstruct it at faster time scales,” said Laura Waller, principal investigator of the study and professor of electrical engineering. and IT. “This is extremely powerful because you can improve your timescales by a factor of 10 or more, depending on how many frames you were initially using.”
NSTM uses machine learning but requires no pre-training or prior data. This simplifies setup and prevents the potential introduction of bias via training data. The only data the model uses are the actual measurements it captured.
In the study, NSTM showed promising results in three different microscopy and photography applications: differential phase contrast microscopy, 3D structured illumination microscopy, and rolling shutter DiffuserCam.
But, according to Waller, “these are really just the tip of the iceberg.” NSTM could potentially be used to improve any multi-shot computational imaging method, thereby expanding its range of scientific applications, particularly in the biological sciences.
“It’s just a model, so you can apply it to any inverse computational problem with dynamic scenes. It could be used in tomography, like CT, MRI, or other super-resolution methods ” she said. “Scanning microscope methods could also benefit from NSTM.”
The researchers envision NSTM one day being integrated into commercially available imaging systems, much like a software upgrade. In the meantime, Cao and others will work to further refine the tool.
“We’re just trying to push the limits of these very fast dynamics,” he said.
More information:
Ruiming Cao et al, Neural space-time model for dynamic multi-shot imaging, Natural methods (2024). DOI: 10.1038/s41592-024-02417-0
Provided by University of California – Berkeley
Quote: Bringing clarity to microscopic imaging: New tool removes motion artifacts (October 1, 2024) retrieved October 1, 2024 from
This document is subject to copyright. Except for fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for informational purposes only.