Credit: Neuron (2024). DOI: 10.1016/j.neuron.2023.12.009
Neurodegenerative diseases are among the most complex human diseases, and their exact causes and mechanisms are the subject of ongoing research and debate. When it comes to Huntington’s disease, the steady accumulation of evidence over the past 30 years has led to a model of molecular events that explains several key features of the disease, including why it appears earlier in some people and why it causes symptoms such as involuntary symptoms. movements and mood swings.
But two new complementary papers from Rockefeller University suggest that may not be the whole story.
Huntington’s disease is caused by somatic expansions of CAGs in which a triple DNA base repeat in a mutated Huntingtin (mHTT) gene increases in number throughout life, leading to cell death. As described in Natural genetics and in Neuron, Rockefeller scientists used a custom technique to reveal that these genetic repeats are unstable and likely produce more toxic proteins, only in certain types of brain cells. Additionally, some cells studied were found to be surprisingly resistant to repeated CAG expansion.
The findings illuminate the cellular nuances of a still-mysterious disease and provide potential targets for therapeutic interventions in the future.
“What we didn’t expect was to see that cells carrying long CAG repeats can survive,” says Nathaniel Heintz, director of the Molecular Biology Laboratory at Rockefeller. “This is very surprising. If expansion is not enough to cause death, what else is needed?”
An awkward threesome
In mHTT, repeated DNA bases (cytosine, adenine, and guanine) form long, unstable stretches, resulting in mutant proteins that damage some neural cells.
The number of CAG repeats closely correlates with disease risk: people with more than 40 repeats will inevitably develop Huntington’s disease, usually between the ages of 30 and 50. The longer the CAG segments, the earlier the symptoms appear. As neurodegeneration snowballs, the disease becomes fatal because we lack treatments that slow or prevent somatic expansion and accumulation of mutant huntingtin.
The molecular processes underlying CAG expansion and cell death are not understood. Over the decades, the Heintz laboratory has generated extensive, high-resolution transcriptional and epigenetic data on the mammalian brain that have, among other things, resulted in the discovery of exciting new neuronal genome modifications that may play critical roles in brain aging and degeneration. More recently, the team decided to apply similar approaches to the study of Huntington’s disease.
“Generating high-quality data from post-mortem samples at early stages of disease can give us insight into what predisposes certain cell types to succumb,” says Heintz.
New methods
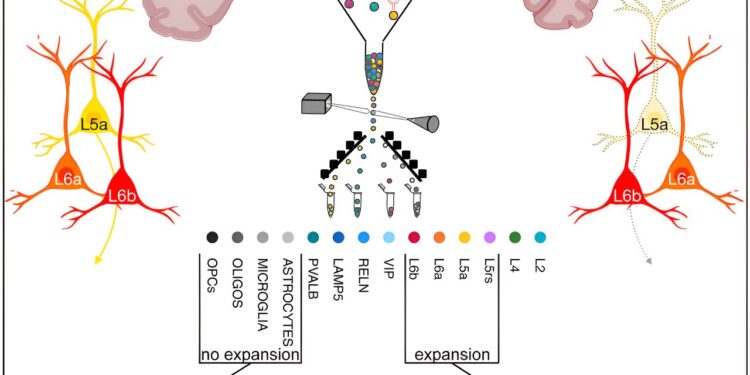
To do this, Kert Mätlik, associate researcher in Heintz’s laboratory and first author of the study Natural genetics article, adapted a cellular analysis method called FANS (fluorescence-activated nuclear sorting) to molecularly profile specific cells in the striatum, a deep brain region connected to motor control and cognition that is greatly affected by Huntington’s disease and Parkinson’s.
By analyzing striatal cells from people who died of Huntington’s disease and donated their brains to science, Mätlik discovered that repeated segments were particularly unstable in medium spiny neurons (MSNs), the most common striatal neurons. These are also the cells that are known to be lost in the striatum during the progression of Huntington’s disease.
Mätlik noticed that the levels of two DNA repair proteins, MSH2 and MSH3, forming the MutSβ complex, were particularly high in MSNs. When cells divide, the role of these proteins is to initiate the repair of incompatible DNA strands, thereby stabilizing the genome by preventing mutations that could otherwise lead to cancer.
Yet when it comes to CAG repeats, these proteins appear to promote CAG expansion rather than prevent it. “Their high levels could really do a disservice to these neurons,” says Mätlik.
Although the resilience of most other striatal cell types can be explained by the stability of CAG repeats, Mätlik was surprised to find that not all cells with CAG expansion are affected in the same way. “This led us to believe that although CAG expansion is a key first step in the pathogenesis of Huntington’s disease, it does not in all cases cause cell death. Another process plays a role,” explains Mätlik.
A new cell type on the map
In the second study, published in Neuronfirst author Christina Pressl, an instructor in clinical investigations at Rockefeller, developed the sFANS (serial fluorescence-activated nuclear sorting) method to isolate different cell types in five regions of the cortex: the motor, cingulate, visual, insular, and prefrontal cortices. . -in 13 early-stage HD brains.
The cortex is made up of six layers, each numbered corresponding to its depth; higher numbers equal deeper layers. The majority of its cells are pyramidal neurons, which use dendrites to sense synaptic inputs and axons to send action potentials, primarily to other parts of the brain. Some of these transmissions travel to the spinal cord.
Pressl used sFANS and deep molecular profiling to identify 16 cell types. “We immediately saw a specific set of cells that was significantly disrupted,” she says.
In the motor region of the cortex, pyramidal cells in layers five and six had extremely long CAG repeats compared to all other cortical cell types. Using sFANS followed by single-cell RNA sequencing, the researchers showed that only cells in layer 5A were more likely to die.
With this discovery, “we put another cell type on the map for increased vulnerability to Huntington’s disease,” notes Pressl.
However, why 5A cells were the least resilient “is really puzzling,” she said. One potential reason is that they project to the striatum, and connectivity between these regions has been shown to weaken in Huntington’s disease. “Perhaps the vulnerable 5A cells found in the cortex are connected to the vulnerable MSN cells found in the striatum,” says Pressl. “When it comes to Huntington’s disease, the entire neural network breaks down at some point.”
Exploring fundamental mysteries
Heintz’s team will pursue several lines of inquiry based on these results. Mätlik will seek to better understand how the MutSβ complex harms rather than cures in SIDS. “Our findings have contributed to the interest in these proteins as potential targets for therapeutic intervention against Huntington’s disease,” notes Mätlik.
Pressl will study the cortex, which is largely unexplored in neurodegenerative diseases: “I am very excited about applying our strategies to multiple cortical regions in the Alzheimer’s brain,” she says.
For Heintz, fundamental questions about the disease remain. “Is there a specific duration of repetitions at which cells become dysfunctional? ” he asks. “If a cell has repeats but doesn’t die, is it dysfunctional enough to cause symptoms? At what CAG repeat length do cells die, and is it different depending on cell type? We need to understand these things in order to develop new cell treatments against this devastating disease.
More information:
Kert Mätlik et al, Cell type-specific CAG repeat expansions and mutant huntingtin toxicity in human striatum and cerebellum, Natural genetics (2024). DOI: 10.1038/s41588-024-01653-6
Christina Pressl et al, Selective vulnerability of layer 5a corticostriatal neurons in Huntington’s disease, Neuron (2024). DOI: 10.1016/j.neuron.2023.12.009
Provided by Rockefeller University
Quote: A new technique for revealing genetic repeats provides surprising information about Huntington’s disease (February 19, 2024) retrieved February 19, 2024 from
This document is subject to copyright. Apart from fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.