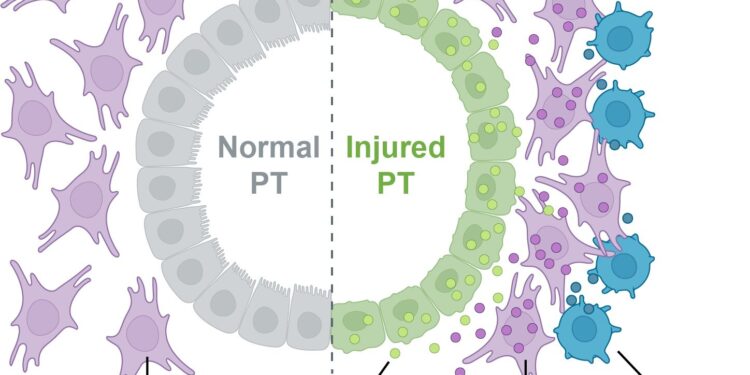
After acute kidney injury, damaged cells interact in disease-promoting microenvironments, a process that can lead to chronic kidney disease. Credit: Michal Polonsky
A study published in Nature Communications This study provides new insights into how damaged cells interact in disease-promoting microenvironments after acute kidney injury, or AKI. Due to limited treatment options, AKI frequently progresses to chronic kidney disease (CKD), which affects more than one in seven American adults, or about 37 million people.
These new findings could help future efforts to prevent CKD, which can lead to kidney failure.
The study brought together scientists from Andy McMahon’s lab at USC and Long Cai’s lab at Caltech.
In the study, co-authors Michal Polonsky of Caltech and Louisa Gerhardt of USC used a cutting-edge tool, called seqFISH, developed in the Cai lab. With this tool, researchers can gather information about gene activity and study cellular interactions in intact kidney tissue in mice with AKI. This allowed the scientists to analyze the precise expression of more than 1,000 genes in injured kidney tissue, identify microenvironments associated with injury, and predict cellular interactions associated with progression to CKD.
“Dr. Cai’s seqFISH technology provides unprecedented insight into cellular interaction in the kidney after injury,” said McMahon, who is the W.M. Keck Dean and University Professor of Stem Cell Biology and Regenerative Medicine at USC and will join the Caltech faculty in October. “A better understanding of kidney injury is needed to identify targets to prevent progression to chronic kidney disease.”
Cai, professor of biology and bioengineering, added: “We are pleased that our technology has led to a better understanding of kidney injury and disease. This study illustrates the importance of interinstitutional and interdisciplinary collaborations to advance biomedical research.”
In the outermost layer of the kidney, the scientists identified a likely pathological microenvironment, which they named “ME-5.” This microenvironment contains a type of kidney cell that is particularly vulnerable to injury, called a proximal tubular cell, or PT.
In ME-5, injured PTs and neighboring connective tissue cells, called fibroblasts, exchange signals that could promote injury progression. Key signals involve the genes Clcf1 and Crfl1, which encode proteins that can promote inflammation and fibrosis, or wound healing. Additional signals detected in ME-5 could contribute to the recruitment of immune cells, further contributing to the development of inflammation, fibrosis, and other pathological changes.
The scientists also identified another important injury-associated microenvironment, which they named “ME-16,” which includes aggregations of various types of immune cells called tertiary lymphoid structures, which are known to contribute to chronic inflammation. Rather than being confined to a specific region of the kidney, ME-16 was distributed throughout the injured organ.
To share their findings, the team constructed a comprehensive map of cellular, molecular, and structural changes following AKI, which refines our understanding of the transition to CKD. This map is publicly available.
Additional co-authors are Kari Koppitch of USC; Jina Yun, Katsuya Lex Colón, Henry Amrhein, Matt Thomson, and Barbara Wold of Caltech; and Shiwei Zheng and Guo-Cheng Yuan of the Icahn School of Medicine at Mount Sinai.
More information:
Spatial transcriptomics defines injury-specific microenvironments and cellular interactions in renal regeneration and disease, Nature Communications (2024). DOI: 10.1038/s41467-024-51186-z
Provided by the Keck School of Medicine of USC
Quote:New study shows cells are involved in unhealthy relationships after acute kidney injury in mice (2024, September 5) retrieved September 5, 2024 from
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without written permission. The content is provided for informational purposes only.