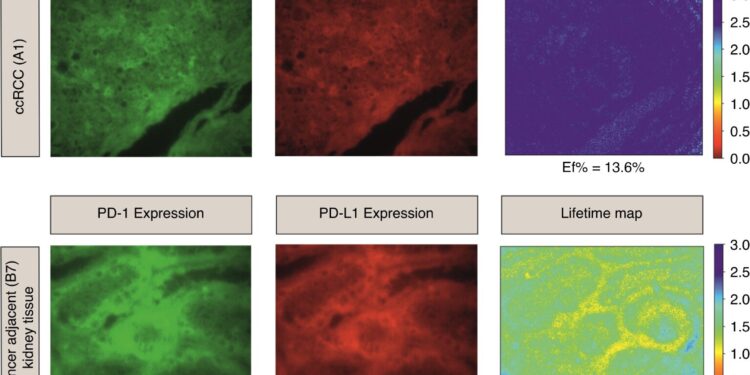
aiFRET quantifies HIF1β/HIF2α and PD-1/PD-L1 interactive states in ccRCC patients. The upper panel illustrates HIF1β/HIF2α expression. HIF1β is the donor and HIF2α is the acceptor. Their lifetime map is presented in the third column with the corresponding calculated median E f. The median E f for normal tissues was 6.70% and for patients A1 and A2 13.6% and 9.30% respectively. The lower panel shows the expression of PD-1 as a donor and PDL1 as an acceptor. PD-1/PD-L1 expression and their average lifespan map with their corresponding calculated median E f are shown. The median Ef for normal tissues was 5.24% and for patients A1 and A2 1.06% and 0% respectively. Credit: BJC Reports (2024). DOI: 10.1038/s44276-023-00033-7
Scientists have developed a new AI tool that maps the function of proteins in a cancer tumor, allowing clinicians to decide how to target treatment more precisely.
In cancers such as clear cell renal cell carcinoma (ccRCC), responses to existing treatments are different for each patient, making it difficult to identify the appropriate drug treatment regimen for each patient. For example, the cancer treatment Belzutifan was recently approved to treat ccRCC, but has only a 49% response rate in patients with the most common form of the disease.
To better understand why some patients respond better than others, researchers from the Universities of Bath and Nottingham studied the function of hypoxia-inducible factor alpha (HIF2α), a key ccRCC target blocked by Belzutifan.
Previous studies have shown that HIF2α levels do not necessarily correlate with tumor aggressiveness and that, counterintuitively, when the levels of the protein present were higher, HIF2α was less active. This means that giving higher doses of Belzutifan potentially exposes the patient to expensive and toxic treatments that may not work and could even make the tumor more resistant to the drugs.
The interdisciplinary team of biophysicists, biologists and computer science researchers designed a new tool, called FuncOmap, which maps the functional state of target oncoproteins on tumor images. This will allow clinicians to directly visualize locations within the tumor where oncoproteins interact, enabling a more accurate diagnosis and indicating the best treatment for each patient. The results are published in the journal BJC Reports.
Diagram illustrating the calculation process. a) Data flow illustrates the progression of information through key stages. b) Interactive FuncOmap in the browser. Credit: BJC Reports (2024). DOI: 10.1038/s44276-023-00033-7
Professor Banafshé Larijani, director of the Center for Therapeutic Innovation at the University of Bath, co-led the study. She said: “People react very differently to medication. It is therefore crucial to be able to predict how patients will respond individually to medications so that a therapy can be tailored to be effective while administering the lowest dose to minimize side effects.
“Our new computational analysis tool uses precision to directly map the functional states of oncoproteins in patients’ tumor sections, so that clinicians can improve patient stratification, thereby enabling personalized medicine.
The team is now collaborating with the laboratory of Dr Amanda Kirane, as well as other surgeons and clinicians at the Stanford University School of Medicine (USA) to further develop and optimize the tool in the field clinical.
Professor Eamonn O’Neill, Head of Bath’s Department of Computer Science and Director of the UKRI Center for Doctoral Training in Responsible, Accountable and Transparent AI (ART-AI), said: “This study describes the type of new and impactful research which is the very essence of interdisciplinary work.
“It brings together computer science, biology and physics, under the umbrella of the UKRI Center for Doctoral Training in Responsible and Transparent Artificial Intelligence, to provide image analysis capable of directly informing clinical decision-making and outcomes Personalized clinics in cancer treatment as well as other diseases.
Professor Jonathan Knight FRS, Vice-President (Business) at the University of Bath, said: “The excitement about this paper lies not only in the work reported, but also in its illustration of how the fields research of biophysics and translational medicine are linked to those of biophysics and translational medicine. Modern computational science promises to accelerate the translation of research into valuable tools for the clinical environment. This really amplifies the value to be gained from transdisciplinary studies.
More information:
Elena Safrygina et al, Spatial functional mapping of hypoxia-inducible factor heterodimerization and immune checkpoint regulators in clear cell renal cell carcinoma, BJC Reports (2024). DOI: 10.1038/s44276-023-00033-7
Provided by the University of Bath
Quote: New spatial tumor mapping tool to assess cancer aggressiveness and personalize treatment (February 9, 2024) retrieved February 9, 2024 from
This document is subject to copyright. Apart from fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.