Credit: Journal of the American Chemical Society (2023). DOI: 10.1021/jacs.3c09195
A team from the University of Massachusetts Amherst has made a major advance in modeling and understanding how intrinsically disordered proteins (IDPs) undergo spontaneous phase separation, an important mechanism of subcellular organization that is at the basis of many biological functions and human diseases.
Displaced people play a crucial role in cancer, neurodegenerative disorders and infectious diseases. They make up about a third of the proteins produced by the human body, and two-thirds of cancer-associated proteins contain large disordered segments or domains. Identifying hidden characteristics essential to the functioning and self-assembly of displaced people will help researchers understand what goes wrong with these characteristics when illnesses arise.
In an article published in the Journal of the American Chemical SocietyLead author Jianhan Chen, professor of chemistry, describes a new way to simulate IDP-mediated phase separations, an important process that has been difficult to study and describe.
“Phase separation is a very well-known phenomenon in polymer physics, but what people didn’t know until about 15 years ago was that it was also a very common phenomenon in biology,” Chen explains. “You can observe phase separation under a microscope, but understanding this phenomenon at the molecular level is very difficult.
“Over the last five or 10 years, people have begun to discover that many of these disordered proteins can cause phase separation, including many important proteins involved in cancer and neurodegenerative disorders.”
The new paper, based on research conducted in Chen’s Computational Biophysics and Biomaterials Laboratory, constitutes a chapter in lead author Yumeng Zhang’s doctoral research. thesis. Zhang will begin work as a postdoctoral researcher at the Massachusetts Institute of Technology (MIT) in February. Another key contributor is Shanlong Li, a postdoctoral research associate in Chen’s lab.
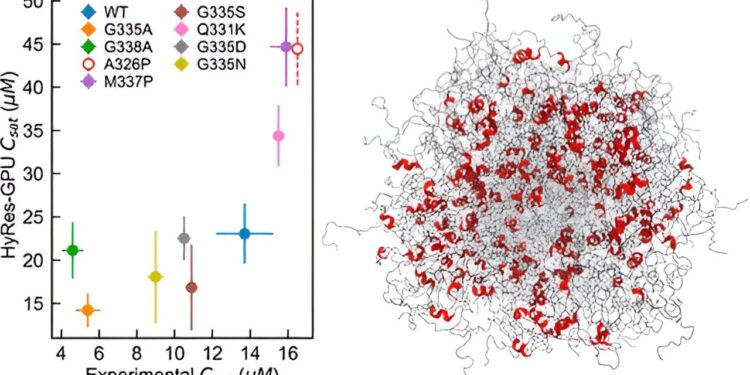
Chen’s lab developed a GPU-accelerated hybrid resolution precise force field (HyRes) to simulate IDP-mediated phase separations. This model is unique in its ability to accurately describe peptide backbone interactions and transient secondary structures, while being computationally efficient enough to model liquid-liquid phase separation. This new model fills a critical gap in existing capabilities for computer simulation of IDP phase separation.
Chen and his team created HyRes simulations to demonstrate for the first time what governs the condensate stability of two important IDPs.
“I actually didn’t expect that it could do such a good job of describing phase separation, because it is a very difficult phenomenon to simulate,” Chen says. “We demonstrated that this model is precise enough to begin to examine the impacts of a single mutation or residual structures in phase separation.”
The researchers’ HyRes-GPU provides an innovative simulation tool to study the molecular mechanisms of phase separation. The ultimate goal is to develop therapeutic strategies in the treatment of diseases associated with disordered proteins.
“That’s really the importance of this work,” Chen says. “Important biological processes are thought to occur through phase separation. So if we can better understand what controls this process, that knowledge will be really powerful, even essential, in allowing us to think about the control of phase separation at various scientific and technical purposes. This will help us understand the type of intervention that will be necessary to achieve therapeutic effects.”
Chen says the next step will be to apply what his team has learned to larger-scale simulations of more complex biomolecular mixtures.
“Shanlong is currently working on building a similar model for nucleic acids, because phase separation often involves both proteins and disordered nucleic acids,” he explains. “We want to be able to describe the two key components, which would allow us to look at many more systems.”
More information:
Yumeng Zhang et al, Towards precise simulation of the coupling between protein secondary structure and phase separation, Journal of the American Chemical Society (2023). DOI: 10.1021/jacs.3c09195
Provided by University of Massachusetts Amherst
Quote: A new simulation tool advances molecular modeling of biomolecular condensates (January 25, 2024) retrieved January 25, 2024 from
This document is subject to copyright. Apart from fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.