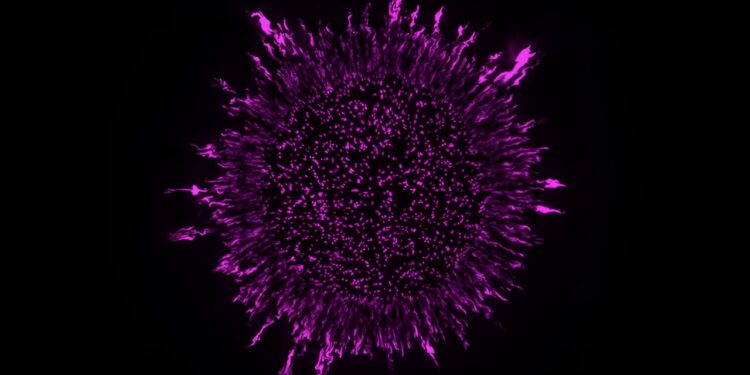
A laboratory-grown Pseudomonas aeruginosa biofilm. The 3D images were acquired by an advanced light-sheet theta microscopy system (ClearScope) developed at Columbia’s Department of Biological Sciences. Credit: Hannah Dayton and Shradha Chauhan, Department of Biological Sciences, Columbia University
Bacteria are traditionally imagined as single-celled organisms, scattered on surfaces or suspended in liquids, but in many environments the true mode of bacterial growth is in sticky clumps called biofilms.
Biofilm formation can be useful to humans; for example, it is an integral part of the production of kombucha tea. But it is most often problematic, because it makes it more difficult to control bacterial growth. When bacterial cells produce a biofilm, it acts as a shield against outside invaders, making the bacteria more tolerant to antibiotics.
Until recently, researchers thought that bacteria were arranged somewhat randomly in biofilms, as much as they had thought about the question of biofilm structure. But new research in the lab of biology professor Lars Dietrich at Columbia University shows that the bacteria that form biofilms actually have a highly structured arrangement within these slimy matrices.
Their unexpected discovery could pave the way for the development of new drugs that better target antibiotic-resistant bacteria.
“There is a yin-yang trade-off for bacteria that form biofilms, since the biofilm protects against antibiotics and other threats, but also prevents food from entering and feeding the system,” said Professor Lars Dietrich, the one of the main authors of the article. “This research gives us an important basis for understanding how to affect the arrangement of bacterial cells and assessing how to make them more sensitive to antibiotics.”
The study, published in the journal Biology PLOS, details research conducted in Professor Dietrich’s laboratory, led by graduate student Hannah Dayton. The article specifically looked at a common and important pathogen called Pseudomonas aeruginosa.
The team used scanning electron microscopy and fluorescence microscopy combined with cell labeling to conduct their research. They found that P. aeruginosa cells in biofilms are packed lengthwise and arranged perpendicular to their growth substrate, the material on which the bacteria live and which contains the substance they consume to survive and thrive. develop. They also found that mutations that change the surface of bacterial cells disrupt this arrangement.
When they tested the effects of an externally added sugar to a fully formed biofilm, they observed that its distribution was affected by the anatomy of the biofilm. Mutant bacteria with a disordered cellular arrangement were more sensitive to added sugar or antibiotics in specific areas of the biofilm, also called subzones. Finally, they showed that changes in biofilm anatomy shift the location of peak metabolic activity within the structure.
Together, these observations indicate that biofilm microstructure is a property that can be tuned to influence the metabolism of resident bacterial subpopulations and affect overall group survival. The results have implications for our approaches to treating infections caused by P. aeruginosa and other biofilm-forming pathogens.
The research was conducted in collaboration with the research groups of Wei Min, professor of chemistry at Columbia; Raju Tomer, professor of biology at Columbia; Jasmine Nirody, professor at the University of Chicago; and Anuradha Janakiraman, professor at the City University of New York (CUNY).
“Cell arrangement is generally an underestimated aspect of biofilm formation,” Dayton said. “We now know that this allows bacteria present in biofilms to control their physiological states and has consequences for their survival during antibiotic treatment.”
“This is a promising development for the pernicious and growing problem of antibiotic-resistant bacteria,” Dietrich said.
More information:
Cellular arrangement impacts metabolic activity and antibiotic tolerance in Pseudomonas aeruginosa biofilms, PLoS Biology (2024). DOI: 10.1371/journal.pbio.3002205
Provided by Columbia University
Quote: New research shows that the arrangement of bacteria in biofilms affects their sensitivity to antibiotics (February 1, 2024) retrieved February 1, 2024 from
This document is subject to copyright. Except for fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.