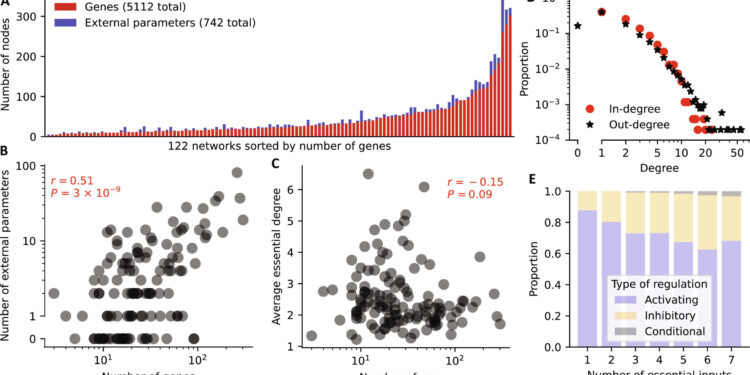
Summary statistics of the analyzed GRN models. (A) Plot of the number of genes and external parameters for each model sorted by number of genes. (B and C) For each model, the number of genes is compared to (B) the number of external parameters and (C) the average essential degree of genes. The Spearman correlation coefficient and associated P value are displayed in red. (D) Distribution of in-degree (red circles) and out-degree (black stars) derived from all 5112 update rules. (E) Prevalence of each regulation type (activation, blue; inhibition, orange; conditional, gray) stratified by the number of regulators (x-axis). Non-essential regulations are excluded. Credit: Scientists progress (2024). DOI: 10.1126/sciadv.adj0822
Over the past 20 years, researchers in biology and medicine have created Boolean network models to simulate complex systems and find solutions, including new treatments for colorectal cancer.
“Boolean network models assume that each gene in a regulatory network can have one of two states: on or off,” explains Claus Kadelka, a systems biologist and associate professor of mathematics at the Iowa State University.
Kadelka and undergraduate student researchers published a study in Scientists progress which unravels common design principles in these mathematical models for genetic regulatory networks.
He says showing which features have evolved over millions of years can “guide the process of building accurate models” for mathematicians, computer scientists and synthetic biologists.
“Evolution has shaped the networks that control our cells’ decision-making in very specific and optimized ways. Synthetic biologists trying to design circuits that perform a particular function can learn from this evolution-inspired design.” , explains Kadelka.
Genetic regulatory networks determine what happens and where it happens in an organism. For example, they cause cells in the lining of your stomach (but not those in your eyes) to produce hydrochloric acid, even though all the cells in your body contain the same DNA.
On a piece of paper, Kadelka draws a simple, hypothetical genetic regulatory network. Gene A produces a protein that turns on gene B, which turns on gene C, which turns off gene A. This negative feedback loop is the same concept as an air conditioner that turns off when a room reaches a certain temperature.
But genetic regulatory networks can be large and complex. One of the Boolean models in the researchers’ dataset involves more than 300 genes. And in addition to negative feedback loops, gene regulatory networks can contain positive feedback loops and feedback loops, which reinforce or delay responses. Redundant genes performing the same function are also common.
Of these and other design principles highlighted in the new paper, Kadelka says one of the most abundant is “channeling.” It refers to a hierarchy or order of importance between genes in a network.
Addison Schmidt (left) and Associate Professor of Mathematics Claus Kadelka (right) stand in front of a blackboard with depictions of a genetic regulatory network in Carver Hall. Credit: Christopher Gannon/Iowa State University
Accessible data, strengthened by undergraduate research
Kadelka points out that the project would have been difficult to complete without the “first-year mentorship program,” which pairs students in Iowa State’s honors program with research opportunities on campus.
The undergraduates helped Kadelka develop an algorithm to analyze 30 million biomedical journal articles and filter out those most likely to include Boolean biological network models. After examining 2,000 articles one by one, the researchers identified around 160 models comprising nearly 7,000 regulated genes.
Addison Schmidt, now a senior in computer science, is one of the paper’s co-authors. When he worked on the project as a freshman in 2021, he created an online database for the project.
“One of the key benefits of the research is that it collects and normalizes Boolean gene regulatory networks from many sources and presents them, along with a set of analysis tools, through a centralized web interface. This expands the accessibility of the data and the web interface makes the analysis tools usable without programming knowledge,” explains Schmidt.
Kadelka says systems biologists have used the database for their research and have expressed gratitude for the resource. He plans to maintain and update the website and study why evolution selects for certain design principles in genetic regulatory networks.
As for Schmidt, he says working on the project as a first-year student helped him broaden his expertise with the Python programming language and become more comfortable applying his skills to research.
“This project also motivated me to pursue further research at Iowa State, where I developed other tools and, coincidentally, another website to showcase them,” Schmidt says.
He adds that he appreciated Kadelka’s mentorship and hopes the first-year mentorship program will continue to foster undergraduate research opportunities at Iowa State.
More information:
Claus Kadelka et al, A meta-analysis of Boolean network models reveals design principles for gene regulatory networks, Scientists progress (2024). DOI: 10.1126/sciadv.adj0822
Provided by Iowa State University
Quote: New research guides the creation of mathematical models for genetic regulatory networks (January 23, 2024) retrieved January 23, 2024 from
This document is subject to copyright. Apart from fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.