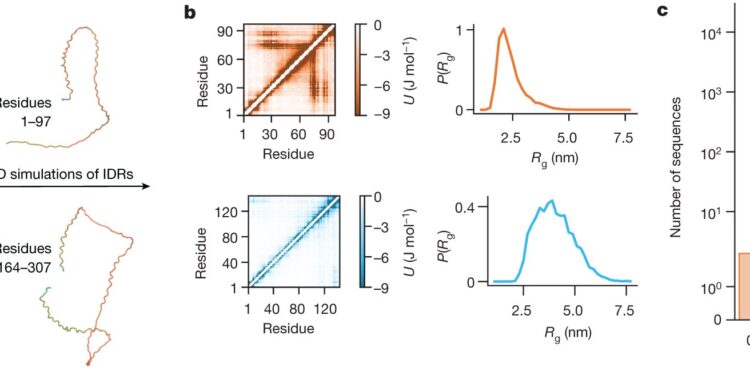
Schematic illustration of the approach used to obtain the conformational properties of all IDRs in the human proteome. a, IDR selection based on the confidence of AlphaFold2 structural prediction, showing selection of two IDRs from nucleosome assembly protein 1-like 3 (NP1L3; UniProt ID: Q99457) as example. b, Molecular dynamics (MD) simulations of the IDRs and calculation of the conformational properties, showing the interaction energy maps and radius of gyration distribution from the simulations of the two IDRs in NP1L3. The simulation data in b are averaged over five independent simulation repeats. c, distribution of the Flory scaling exponent, ν, for 28,058 IDR in the human proteome; note the logarithmic scale. Some IDRs have very small values of ν (0.3% have ν < 0.33); these IDRs show conformational properties that are not well described by a simple scaling law. We note that we do not find evidence of significant enrichment of misclassified structured sequences in compact IDRs. Credit: Nature (2024). DOI: 10.1038/s41586-023-07004-5
Protein molecules are at the heart of biology. Our typical understanding of proteins states that each type of protein has a specific three-dimensional shape that allows it to perform its function. This dogma is challenged by intrinsically disordered proteins, which make up a third of all proteins and have central biological functions even though their shapes are constantly changing.
Until now, our understanding of the structural properties of this intriguing class of proteins has relied on the study of only a small number of examples. In research published today in the journal NatureResearchers from the Department of Biology at the University of Copenhagen have shown how all the disordered proteins (around 28,000) in the human body behave.
“I have always been fascinated by intrinsically disordered proteins because they seem to defy most of the rules governing how a protein behaves. Over the past 20 years, we have worked to determine what these strange proteins look like and whether new rules had to be applied. applied to describe them. For the first time, we were able to study the structure of all disordered human proteins and began to bring order to this world of molecular disorder,” says Professor Kresten Lindorff -Larsen, director of the NNF PRISM center, in which the research was carried out.
The goal of the PRISM center is to combine computational methods from biophysics and machine learning with methods from cell biology to study how genetic variants cause disease. But until now, researchers didn’t know what most disordered proteins looked like, and so couldn’t even begin to study how mutations in the genes encoding them might cause disease.
Until recently, we looked at disordered proteins one by one, and it was essential to find a way to study them on a larger scale,” says Assistant Professor Giulio Tesei, one of the lead authors of the new paper. We proposed an approach in which we could use experimental measurements on disordered proteins to develop a computational model to predict their properties. Since this model is both accurate and fast, we can now examine them all. »
The study was co-led by undergraduate student Anna Ida Trolle, who said: “When I started the project, I didn’t know that we usually study one or two proteins at a time. So when Giulio and Kresten suggested that I should study some 28,000 proteins, luckily I didn’t realize what a crazy idea it was, but we quickly found a way to generate and track a large amount of data and were able to use it to study biology and evolution. of disordered proteins.
Lindorff-Larsen concludes: “This has been a challenging but also extremely fun project, which was only made possible thanks to the contributions of several people with diverse expertise within the PRISM center. We have taken new steps in linking the molecular properties of disordered proteins. to their biological function and their roles in disease. Finally, we begin to understand the language of disordered proteins.
More information:
Giulio Tesei et al, Conformational ensembles of the intrinsically disordered human proteome, Nature (2024). DOI: 10.1038/s41586-023-07004-5
Provided by the University of Copenhagen
Quote: New research brings order to disordered proteins (January 31, 2024) retrieved January 31, 2024 from
This document is subject to copyright. Except for fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.