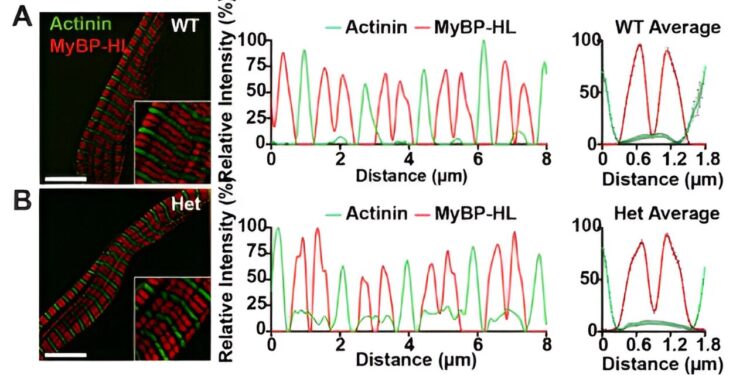
Structured illumination microscopy of atrial cardiomyocytes isolated from wild-type (A) and heterozygous Mybphl-null mice (B) reveals localization of MyBP-HL (red) in an Aband doublet pattern interdigitated with actinin-stained Z disks (green) in heterozygous wild-type and Mybphl mice. Credit: Proceedings of the National Academy of Sciences (2023). DOI: 10.1073/pnas.2314920120
Investigators led by Elizabeth McNally, M.D., Ph.D., Elizabeth J. Ward Professor of Genetic Medicine and director of the Center for Genetic Medicine, have discovered previously unknown protein interactions in the atrium of the heart that are essential for normal cardiac function, according to findings published in the Proceedings of the National Academy of Sciences.
The heart is divided into four chambers: two at the top (atria/atria) and two at the bottom (ventricles), with one on each side of the heart. The ability of the ventricles of the heart to pump oxygen-poor blood to the lungs and oxygen-rich blood to the rest of the body depends on the atria, which constantly fill the ventricles with blood.
The atria are also a primary site of disease, including atrial fibrillation, which occurs when the atria no longer contract normally and instead have rapid, uncoordinated, irregular heartbeats, or arrhythmia. These chaotic beats alter the normal filling of the ventricles and can eventually lead to a stroke or even heart failure.
“We are finally starting to have good tools to look at the atria and recognize the relaxation properties as important and essential for overall heart function. If the atria cannot fully do their job, then the ventricles cannot. do. their complete job,” said McNally, who is also a professor of medicine in the Division of Cardiology and Biochemistry and Molecular Genetics.
However, the precise mechanisms regulating atrial contraction and relaxation have recently attracted more attention with the discovery of new, previously unknown proteins, according to McNally.
In a previous study from the McNally laboratory published in Traffic, investigators discovered the MYPBHL gene and that mutations in this gene increase the risk of arrhythmia and cardiomyopathy. MYBPHL is a gene that encodes the myosin-binding protein H-like (MyBP-HL), which is part of the contractile machinery found primarily in the atria.
Additionally, MyBP-HL belongs to the same family of proteins as myosin-binding protein C (cMyBP-C), which acts as a braking system for the heart to prevent it from contracting excessively. Mutations in the gene encoding cMyBP-C are an important cause of hypertrophic cardiomyopathy. However, the relationship between these two proteins and their combined impact on the function of the ventricles and atria remains poorly understood.
In the present study, using structured illumination microscopy, immunoelectron microscopy and mass spectrometry to analyze cardiac cells from genetic mouse models, the team identified a novel binding relationship between MyBP-HL and cMyBP- vs.
Specifically, loss of MyBP-HL doubled the amount of cMyBP-C in the atria, while loss of cMyBP-C doubled the amount of MyBP-HL in the atria. Loss of MyBP-HL also accelerated atrial relaxation.
Overall, the results highlight a novel mechanism and an essential role of MyBP-HL in the regulation of relaxation and atrial function.
The findings could also shed light on abnormal atrial relaxation properties seen in heart failure and in the heart as it ages, according to McNally. The heart, particularly the atria, becomes stiffer with age, and MyBP-HL may also be a biomarker of atrial abnormalities, such as atrial fibrillation, according to the authors.
“Our work on MYBPHL has sparked interest in how contraction of the atria differs from that of the ventricles. These results are exciting because we now have a potential new therapeutic target to modulate atrial contractile function,” said Dave Barefield , Ph.D., former postdoctoral fellow in the McNally lab and first author of the study.
More information:
David Y. Barefield et al, Myosin-binding protein H regulates the distribution and function of myosin-binding proteins in atrial cardiomyocytes, Proceedings of the National Academy of Sciences (2023). DOI: 10.1073/pnas.2314920120
Provided by Northwestern University
Quote: Novel protein interactions may serve as a biomarker for heart disease (January 17, 2024) retrieved January 17, 2024 from
This document is subject to copyright. Except for fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.