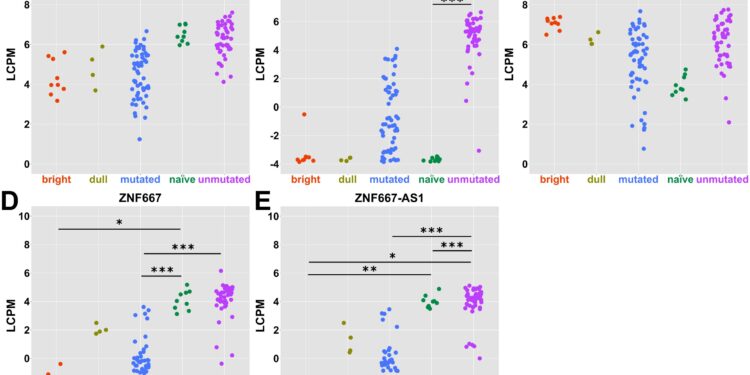
Traditional and potential biomarkers for CLL subtypes. Credit: PLOS One (2025). DOI: 10.1371/journal.pone.0335069
Researchers from the University of Eastern Finland and their international collaborators have identified key developmental and molecular differences between the two main subtypes of chronic lymphocytic leukemia, CLL. The results, published in PLOS ONEshow that mutated and non-mutated forms of CLL may arise from distinct stages of B cell development, providing new insights into disease mechanisms and biomarker discovery.
Mutated and non-mutated LLCs take different developmental pathways
CLL, the most common leukemia in adults, is characterized by disruption of the peripheral immune system through the accumulation of abnormal B lymphocytes. CLL is divided into mutated (M-CLL) and non-mutated (UM-CLL) subtypes based on the frequency of mutation of the immunoglobulin heavy chain variable region in B cells. UM-CLL is more aggressive and tends to have a worse prognosis than M-CLL. The research team performed a meta-analysis of transcriptomic data from 116 patients and B cells from healthy donors to explore the origins of these subtypes.
B cells go through different stages of development in the bone marrow and in the germinal centers of lymphatic tissue. They are classified into different subtypes based on their maturation and function, such as memory cells or plasma cells. Results revealed that M-CLL resembles the germinal center-dependent memory B cell subtype, CD27bright memory B cells, while UM-CLL reflects an earlier intermediate germinal center stage, possibly explaining their differences in mutation levels and clinical behavior.
New molecular biomarkers identified
The study highlights three genes – LPL, ZNF667 and ZNF667-AS1 – as promising biomarkers for more accurate stratification of CLL patients. These genes are involved in cholesterol regulation and epithelial-mesenchymal transition, processes that may contribute to disease progression and aggressiveness.
“Our results suggest that the two CLL subtypes may trace back to B cells in the germinal center but diverge at different points of development,” says lead author Ahmed Mohamed from the University of Eastern Finland. “This helps us better understand why some patients have more aggressive disease than others.”
“The identification of genes such as LPL and ZNF667 gives us tools for more precise molecular classification and potentially for new therapeutic strategies,” adds Dr. Ola Grimsholm from the Medical University of Vienna.
Shared and distinct paths
Furthermore, compared to healthy B cells, both CLL subtypes showed alterations in pathways related to neuroactive ligand-receptor signaling and cell-cell adhesion, while subtype-specific differences involved cholesterol metabolism (UM-CLL) and immune activation and platelet signaling (M-CLL). These findings strengthen the link between metabolic regulation, immune signaling and leukemia biology.
More information:
Ahmed Mohamed et al, Germinal center trajectories and transcriptional signatures define CLL subtypes and their pathway regulators, PLOS One (2025). DOI: 10.1371/journal.pone.0335069. journals.plos.org/plosone/arti …journal.pone.0335069
Provided by the University of Eastern Finland
Quote: Chronic lymphocytic leukemia: new origins and biomarkers revealed (November 18, 2025) retrieved November 18, 2025 from
This document is subject to copyright. Except for fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for informational purposes only.