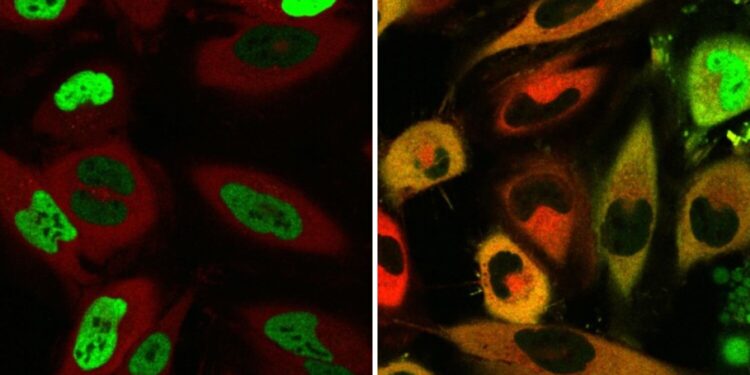
Cells before and after TRAM introduction. TRAMs bind a shuttle protein (red) and a target protein (green). Without TRAM, the target protein resides in the nucleus (left), and upon TRAM treatment, the target protein is pulled into the cytoplasm by the shuttle protein (right). Credit: Steven Banik and Christine Ng
Cells are highly controlled spaces that depend on every protein being in the right place. Many diseases, including cancers and neurodegenerative disorders, are associated with misplaced proteins. In some cancers, for example, a protein that normally monitors DNA replication in the nucleus is sent far from the DNA it is supposed to monitor, allowing cancers to grow.
Steven Banik, an assistant professor of chemistry in the School of Humanities and Information Sciences and a researcher at Stanford University’s Sarafan ChEM-H Institute, and his lab have developed a new method to help force displaced proteins back into their proper place in cells. The method involves rewiring the activity of natural shuttles to help move proteins to different parts of the cell. The team designed a new class of molecules called “targeted relocation activating molecules,” or TRAMs, that convince these natural shuttles to take a different cargo with them, such as proteins exported from the nucleus in some cancers. Published in Nature On September 18, this strategy could lead to a therapy to correct the misplacement of proteins associated with diseases, and also to create new functions in cells.
“We recover lost proteins and bring them home,” Banik said.
Shuttles and passengers
Our cells contain many compartments, such as the nucleus, the safe haven for DNA, or the mitochondria, where energy is produced. Between all these compartments is the cytoplasm. Throughout the many places in the cell are proteins. They are responsible for all sorts of actions (building and breaking down molecules, contracting muscles, sending signals), but to function properly, they must perform their respective actions in the right place.
“Cells are busy places,” Banik says. “Proteins weave through the crowd, past all sorts of other molecules like RNA, lipids, and other proteins. So a protein’s function is limited by what it can do and how close it is to other molecules.”
Diseases sometimes exploit this need for proximity by mutating proteins that might otherwise protect a cell from damage. This type of mutation is like putting the wrong address on a package, prompting proteins to go where they would never go in healthy cells.
Sometimes this movement stops the protein from working altogether. Proteins that act on DNA, for example, can’t find DNA in the cytoplasm and fly away without doing anything. In other cases, this movement causes a protein to become a bad actor. In ALS, for example, a mutation sends a certain protein, called FUS, out of the nucleus and into the cytoplasm, where it clumps together in toxic clumps and eventually kills the cell.
Banik and his team wondered if they could combat this intentional movement of proteins by using other proteins as shuttles to carry the passenger proteins to their destination. But these shuttles often have other functions, so the team would have to convince the shuttle to take the cargo and transport it to a new location.
To do this, Banik and his team developed a new type of two-headed molecule called TRAM. One head is designed to stick to the shuttle, and the other to stick to the passenger. If the shuttle is strong enough, it will carry the passenger in its place.
On the way to the trip
The team focused on two promising types of shuttles, one that shuttles proteins into the nucleus, and one that exports proteins from the nucleus. Christine Ng, a graduate student in chemistry and first author of the paper, designed and built TRAMs that connect the shuttle and the passenger. If a passenger from the cytoplasm ends up in the nucleus, they will know their TRAM has worked.
The first challenge was immediate: There was no reliable way to measure the amount of a protein at a specific location in individual cells. So Ng developed a new method to quantify the amount and location of passenger proteins in a cell at a given time. A chemist by training, she had to learn new skills in microscopy and computational analysis to do this.
“Nature is inherently complex and interconnected, so it’s essential to adopt interdisciplinary approaches,” Ng said. “Borrowing logic or tools from one field to solve a problem in another often leads to very interesting questions and discoveries.”
She then put the system to the test. Her TRAMs successfully moved passenger proteins in and out of the nucleus, depending on which shuttle was being used. These early experiments helped her generate some basic “rules” for design, such as how much resistance a shuttle needed to have to overcome the passenger’s tendency to pull in another direction.
The next challenge was to design TRAMs that could be drugs—that is, reverse the movement of disease-causing proteins. They first created a TRAM that would relocalize FUS, the protein that gets kicked out of the nucleus and forms dangerous granules in ALS patients. After treating cells with their TRAM, the team found that FUS was transported back to its original location, the nucleus, the toxic clumps shrank, and the cells were less likely to die.
They then turned their attention to a well-known mutation in mice that makes them more resistant to neurodegeneration. This mutation, studied in particular by Ben Barres, now deceased, causes the displacement of a certain protein from the nucleus to the axon of neurons.
The team wondered if it was possible to create a TRAM that would mimic the protective effect of the mutation, by carrying the protein to the end of the axon. Their TRAM not only moved the target protein along the axon, but also made the cell more resistant to the stress that mimics neurodegeneration.
In all of these examples, the team faced an ongoing challenge: Designing the TRAM head to target passengers is difficult, because scientists have not yet identified all of the molecules that might bind to their target passengers. To get around this problem, the team used genetic tools to install a sticky tag on these passengers. In the future, however, they hope to find naturally occurring sticky bits on these passengers and develop TRAMs into new types of drugs.
Although they focused on two shuttles, the method is generalizable to all other shuttles, such as those that push objects to the cell surface, where communication with other cells occurs.
And beyond sending mutated proteins back where they belong, the team also hopes that TRAMs could be used to send healthy proteins to parts of the cell they can’t normally reach, creating new functions that we don’t yet know are possible.
“It’s exciting because we’re just beginning to understand the rules,” Banik said. “If we shift the balance, if a protein suddenly has access to new molecules in a new part of the cell at a different time, what will it do? What functions could we unlock? What new part of biology could we understand?”
More information:
Christine SC Ng et al, Targeted protein relocation via protein transport coupling, Nature (2024). DOI: 10.1038/s41586-024-07950-8
Provided by Stanford University
Quote:New method developed to relocate misplaced proteins in cells (2024, September 21) retrieved September 22, 2024 from
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without written permission. The content is provided for informational purposes only.