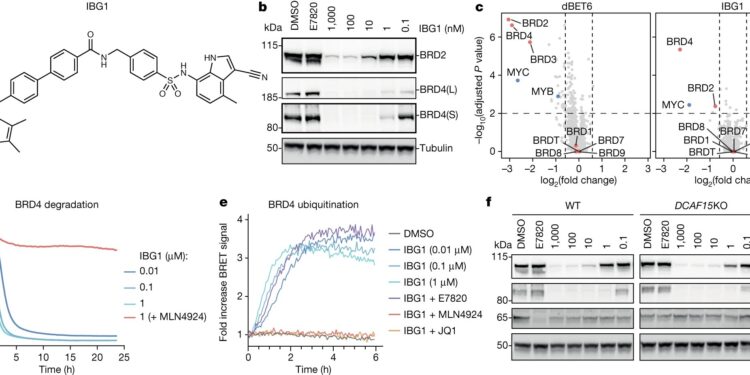
IBG1 degrades BRD2 and BRD4 independently of DCAF15. AStructure of IBG1. b, BET protein degradation activity of IBG1. HEK293 cells were treated for 6 h with DMSO, E7820 (1 μM), or increasing concentrations of IBG1. BET protein was quantified by immunoblotting. Data representative of not = 3 independent experiments. vs, Whole proteome changes after degradative treatment. Quantitative proteomics in KBM7 cells was performed after 6 h of treatment with DMSO, IBG1 (1 nM), or dBET6 (10 nM). save2-transformed fold change and −logten-adjusted Benjamini–Hochberg transformed one-way analysis of variance (ANOVA) P.value compared to DMSO treatment. not = 3 biological repetitions. d, NanoBRET kinetic degradation assay. BromoTag–HiBiT–BRD4 knock-in HEK293 cells were treated with IBG1 with or without MLN4924 pretreatment (10 µM) for 1 h. Average of not= 3 biological repetitions. RLU, relative light units. e, NanoBRET kinetic ubiquitination assay. LgBiT-transfected HEK293 HiBiT–BromoTag–BRD4 knock-in cells were treated with IBG1 at the indicated concentrations or at 10 nM after pretreatment with JQ1, E7820 (both 10 µM), or MLN4924 (1 µM) for 1 h. Average of not = 4 biological repetitions. F, DCAF15-independent BET protein degradation. Wild type (WT) and DCAF15HCT-116 knockout (KO) cells were treated with increasing concentrations of IBG1 for 6 h and BET protein was quantified by immunoblotting. Data representative of not= 3 independent experiments. Credit: Nature(2024). DOI: 10.1038/s41586-024-07089-6
A revolutionary class of molecular glue identified at the University of Dundee could pave the way for a new generation of drugs targeting cancers and neurodegenerative diseases.
A research team from the University Center for Targeted Protein Degradation (CeTPD) led by Professor Alessio Ciulli, in collaboration with the research group of Dr. Georg Winter at the Research Center for Molecular Medicine (CEMM) of the Austrian Academy of Sciences in Vienna, defined a new class of “intramolecular bivalent glue,” which binds proteins – crucial to the cells that allow our bodies to function properly – that would otherwise remain separated.
This research was published in the journal Nature.
“These results have major implications for the entire pharmaceutical industry engaged in the search for targeted protein degraders,” said Professor Alessio Ciulli, Director of CeTPD in Dundee.
“This is particularly true for the development of drugs targeting cancer, neurodegenerative diseases, and many other diseases caused by proteins that have historically been considered non-druggable.”
“Protein is essential for our cells to function properly, but when cells do not function properly, the body is vulnerable to disease.”
“The glue we were able to define is special because it first attaches to a protein in two places (not just one) and then recruits the second protein, thereby sandwiching them.”
“We were only able to identify this using our targeted protein degradation technology and have identified a vulnerability that can be exploited by the design of new drugs that could transform the treatment of patients with cancer and those with other incurable diseases.”
Targeted protein degradation (TPD) is an emerging area of drug development to treat disease that involves repurposing protein recycling systems in our cells to destroy disease-causing proteins. Most TPD strategies use small molecules, called degraders, to recruit these target proteins to a class of enzymes called ubiquitin E3 ligases.
E3 labels the target protein with ubiquitin markers, which ultimately leads to the destruction of the pathogenic protein via the cellular trash can: the proteasome.
Working with collaborators from CEMM, Goethe University Frankfurt and Eisai Co. Ltd, the Japanese pharmaceutical company, the Dundee team was able to unveil a new molecular bonding mechanism, different from those previously known. This new mechanism binds to both sides of the target protein instead of just one, causing a rearrangement of the entire protein and stabilizing its previously unknown interaction with E3 ligase.
Additionally, the team was able to visualize, for the first time, the precise mechanism by which their compounds act and bring together target proteins to one of these E3 ligases. Because the molecules have two heads, which attach to two different regions within the same target protein, they have been called “intramolecular bivalent glues.”
This cutting-edge work has also shed light on previously underestimated characteristics and properties of molecular glues, paving the way for scientists to develop a deeper understanding of glues, which could lead to new classes being discovered more quickly.
“The impact of what we have revealed here cannot be underestimated,” added Professor Ciulli. “This will have a ripple effect throughout the pharmaceutical industry and could potentially transform the way we view drug development.” I must also pay tribute to our collaborators, whose contribution was crucial in achieving this seismic breakthrough.
More information:
Alessio Ciulli, Targeted protein degradation via intramolecular bivalent genes, Nature(2024). DOI: 10.1038/s41586-024-07089-6. www.nature.com/articles/s41586-024-07089-6
Provided by the University of Dundee
Quote: New class of “intramolecular bivalent glue” could transform cancer drug discovery (February 21, 2024) retrieved February 21, 2024 from
This document is subject to copyright. Apart from fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.