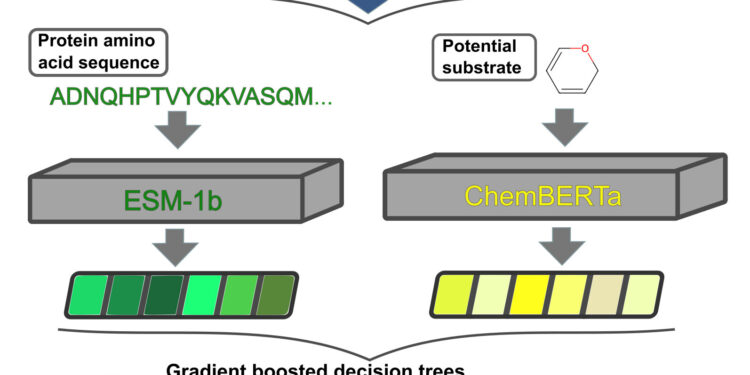
Schematic diagram of the prediction process: Protein Databases provides a dataset containing 8,500 experimentally validated transporter-substrate pairs to train the model (top). Transport proteins comprise a sequence of amino acids converted into vectors by a deep learning model (center left, in different shades of green). Information about potential substrates is also converted into digital vectors (center right, in different shades of yellow). These vectors train a so-called gradient boosting model (set of multiple decision trees) to predict whether the molecule is a substrate for a specific transport protein (bottom). Credit: HHU/Alexander Kroll
Transport proteins are responsible for the continuous movement of substrates into and out of a biological cell. However, it is difficult to determine which substrates a specific protein can transport. Bioinformaticians at Heinrich Heine University Düsseldorf (HHU) have developed a model called SPOT that can predict this with a high degree of accuracy using artificial intelligence (AI).
The researchers present their approach, which can be used with arbitrary transport proteins, in the journal Biology PLOS.
Biological cell substrates must be continually transported in and out across the cell membrane to ensure cell survival and enable them to perform their function. However, not all substrates that move through the body should be able to enter cells. And some of these transport processes must be controllable so that they only occur at a certain time or under specific conditions in order to trigger cellular function.
The role of these active and specialized transport channels is assumed by so-called transport proteins (transporters), a wide variety of which are integrated into cell membranes. A transport protein consists of a large number of individual amino acids that together form a complex three-dimensional structure.
Each transporter is tailored to a specific molecule, called a substrate, or a small group of substrates. But which one, exactly? Researchers are constantly searching for matching transporter-substrate pairs.
Professor Martin Lercher, from the Computational Cell Biology Research Group and corresponding author of the study, says: “It is difficult to determine experimentally which substrates correspond to which transporters. Even determining the three-dimensional structure of a transporter – from which it may be possible to identify substrates – is challenging, because proteins become unstable as soon as they are isolated from the cell membrane.
“We chose a different approach, based on AI,” explains Dr. Alexander Kroll, lead author of the study and postdoctoral researcher in Professor Lercher’s research group. “Our method, called SPOT, used more than 8,500 transporter-substrate pairs, which have already been validated experimentally, as a training dataset for a deep learning model.”
To enable a computer to process transport proteins and substrate molecules, Düsseldorf bioinformaticians first convert the protein sequences and substrate molecules into digital vectors, which can be processed by AI models. Once the learning process is complete, the vector of a new transporter and that of potentially suitable substrates can be input into the AI system. The model then predicts the probability that certain substrates match the transporter.
Kroll explains: “We validated our trained model using an independent test dataset in which we already knew the transporter-substrate pairs. SPOT predicts with greater than 92% accuracy whether an arbitrary molecule is a substrate for a specific transporter. »
SPOT thus suggests very promising substrate candidates. “This allows us to significantly limit the search scope of experimenters, speeding up the process of identifying the substrate that definitively corresponds to a transporter in the laboratory,” says Professor Lercher, explaining the link between bioinformatics prediction and experimental verification.
Kroll adds: “And this applies to any arbitrary transport protein, not just limited classes of similar proteins, as is the case in other approaches to date.”
There are different potential application areas for the model.
Lercher notes: “In biotechnology, metabolic pathways can be modified to enable the manufacture of specific products such as biofuels, or drugs can be tailored to transporters to facilitate their entry into precisely the cells in which they are intended to have an effect. »
More information:
Alexander Kroll et al, SPOT: A machine learning model that predicts specific substrates for transport proteins, Biology PLOS (2024). DOI: 10.1371/journal.pbio.3002807
Provided by Heinrich-Heine-University Düsseldorf
Quote: New AI model can predict movement of substrate into and out of cells (September 26, 2024) retrieved September 26, 2024 from
This document is subject to copyright. Except for fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for informational purposes only.