Credit: Cell (2024). DOI: 10.1016/j.cell.2024.08.038
“Gut health” is an increasingly common buzzword among foodies and dietitians, and for good reason. The billions of microbes and bacteria living in our gut are involved in many aspects of health and disease.
Yale Microbial Sciences Institute researchers have taken an important step toward personalized, evidence-based nutrition tailored to individual gut health needs.
Andrew Goodman’s lab has generated the first systematic map showing how molecules in certain foods interact with our unique gut bacteria. Their findings are published in the journal Cell.
Building on previous research examining drugs and gut bacteria, scientists sought to understand why different people react differently to the same foods.
“We know that diet is an essential part of our health and shapes our microbiome,” explained Elizabeth Culp, a former postdoctoral fellow at the Goodman Lab and first author of the study.
While a large body of work has described the effects of “macronutrients” such as fiber on our gut microbiomes, surprisingly little is known about how other small molecules in foods cause health problems.
“Aside from anecdotal examples in the scientific literature, evidence is scant regarding dietary changes people can make to help them manage risk factors for diseases like diabetes or cancer,” Culp said.
“It’s possible that this is because our microbiomes react differently to the same molecules found in foods.”
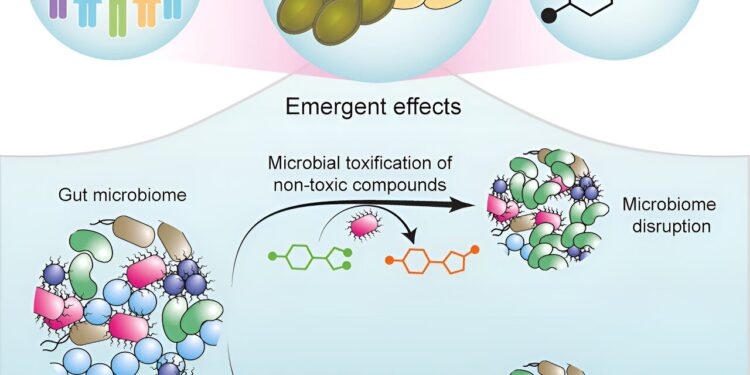
Researchers have designed a systematic map of the interactions between small molecules present in our foods and the different bacteria present in the intestine.
This work is among the first to describe the specific microbial genes responsible for the metabolic transformation of food compounds and the mechanisms by which food compounds modify our microbiomes.
Using liquid chromatography and mass spectrometry at the Yale West Campus Analytical Core, scientists combined different molecules with gut bacteria to create growth models and maps for approximately 150 “xenobiotic” food compounds. Sequencing at the Yale Center for Genome Analysis allowed the team to measure the degree of change in the composition of human gut communities.
“We were surprised by the level of variability,” said Goodman, the CNH Long Professor and chair of microbial pathogenesis, and director of the Microbial Sciences Institute (MSI).
“The same dietary compound could significantly reshape some people’s gut microbial communities while having virtually no impact on other people’s microbiomes,”
The molecular map provides a mechanism to explain these variable responses between different people, showing how a dietary compound affects the growth of gut microbes and how that compound is metabolically modified by the microbial community.
It remains difficult to predict how an individual reacts to a given food and, ultimately, how it affects their health. But the findings provide a basis for understanding how metabolic reactions vary from person to person and how these differences shape the growth of “good” or “bad” bacteria in our gut.
“If we can understand the specific microbial genes that determine how a microbiome responds to a molecule in our food, and how these genes are different between different people’s microbiomes, the correlations with diseases like cancer, diabetes or gastrointestinal infections can start to make sense,” concludes Culp, who is currently a scientist at Empress Therapeutics in Boston.
“This is the first step toward creating personalized dietary recommendations as part of personalized nutrition strategies.”
More information:
Elizabeth J. Culp et al, Microbial transformation of dietary xenobiotics shapes the composition of the gut microbiome, Cell (2024). DOI: 10.1016/j.cell.2024.08.038
Cell
Provided by Yale University
Quote: Molecular map shows the way to better food choices (October 4, 2024) retrieved October 4, 2024 from
This document is subject to copyright. Except for fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for informational purposes only.