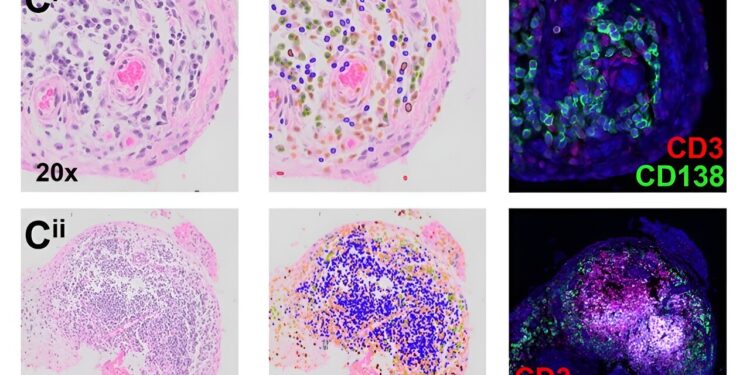
A machine learning model can predict with high accuracy lymphocyte cells, shown in the blue and green overlays in the middle panel, on H&E histology slides. The predictions are confirmed by immunofluorescent staining of B cells (CD138 or CD20) and T cells (CD3). Credit: Adapted from Nature Communications (2024). DOI: 10.1038/s41467-024-51012-6
A machine learning tool created by researchers at Weill Cornell Medicine and Hospital for Special Surgery (HSS) can help distinguish between subtypes of rheumatoid arthritis (RA), which may help scientists find ways to improve care for this complex disease.
The study, published in Nature Communicationsshows that artificial intelligence and machine learning technologies can effectively and efficiently subtype pathology samples from RA patients.
“Our tool automates the analysis of pathology slides, which could one day lead to more accurate and effective disease diagnosis and personalized treatment of RA,” said Fei Wang, MD, professor of population health sciences and founding director of the AI Institute for Digital Health (AIDH) in the Department of Population Health Sciences at Weill Cornell Medicine. “It shows that machine learning has the potential to transform pathology assessment for many diseases.”
There has already been some research on machine learning tools to automatically analyze pathology slides in oncology. Dr. Wang and his colleagues have been working to expand the use of this technology to other clinical specialties.
Automate a slow process
For the latest study, Dr. Wang teamed up with Dr. Richard Bell, an instructor in the Arthritis and Tissue Degeneration Program and Research Institute and a computational pathology analyst in the HSS Molecular Histopathology Core Laboratory, and Dr. Lionel Ivashkiv, scientific director and chair of the Arthritis and Tissue Degeneration Program at HSS and professor of medicine at Weill Cornell Medicine, to automate the process of subtyping RA tissue samples. Distinguishing between the three RA subtypes can help clinicians choose the treatment most likely to be effective for a particular patient.
Pathologists currently manually classify arthritis subtypes using a rubric to identify cellular and tissue features in biopsy samples from human patients, a slow process that increases the cost of research and can lead to inconsistencies among pathologists.
“This is the analytical bottleneck of pathology research,” Dr. Bell said. “It’s a very time-consuming and tedious task.”
The team first trained their algorithm on RA samples from one group of mice, optimizing its ability to distinguish tissue and cell types in the sample and sort them by subtype. They then validated the tool on a second group of samples. The tool also provided new insights into treatment effects in mice, such as reduced cartilage breakdown within six weeks of receiving commonly used RA treatments.
They then deployed the tool on patient biopsy samples from the Accelerating Medicines Partnership Rheumatoid Arthritis Research Consortium and showed that it could effectively type human clinical samples. The researchers are now validating the tool with additional patient samples and determining how best to integrate this new tool into pathologists’ workflows.
A step towards personalized medicine
“This is the first step toward more personalized care for RA,” Dr. Bell said. “If you can create an algorithm that identifies a patient’s subtype, you can get them the treatments they need more quickly.”
The technology could provide new insights into the disease by detecting unexpected tissue changes that humans might miss. By saving pathologists time on subtyping, the tool could also reduce the cost and increase the efficiency of clinical trials testing treatments for patients with different RA subtypes.
“By integrating pathology slides with clinical information, this tool demonstrates the growing impact of AI in advancing personalized medicine,” said Rainu Kaushal, MD, senior associate dean of clinical research and chair of the Department of Population Health Sciences at Weill Cornell Medicine. “This research is particularly exciting because it opens new avenues for detection and treatment, making significant advances in how we understand and care for people with rheumatoid arthritis.”
The team is currently working on developing similar tools to assess osteoarthritis, disc degeneration, and tendinopathy. In addition, Dr. Wang’s team is also looking to define disease subtypes from broader biomedical information. For example, they recently demonstrated that machine learning can distinguish three subtypes of Parkinson’s disease.
“We hope our research will inspire more computational research into developing machine learning tools for more diseases,” Dr. Wang said.
“This work represents an important advance in RA tissue analysis that can be applied to benefit patients,” added Dr. Ivashkiv.
More information:
Richard D. Bell et al, Automated multiscale computational pathotyping (AMSCP) of inflamed synovial tissue, Nature Communications (2024). DOI: 10.1038/s41467-024-51012-6
Provided by Weill Cornell Medical College
Quote: Machine learning helps identify subtypes of rheumatoid arthritis (2024, August 29) retrieved August 29, 2024 from
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without written permission. The content is provided for informational purposes only.