A sensor platform that regulates bacterial gene expression via targeted substances using diverse aptamer sequences. Credit: POSTECH
A research team has developed an RNA-based sensor platform capable of regulating gene expression in bacteria. Their findings were recently presented in the journal Advanced science.
The concept of managing obesity by stimulating beneficial microorganisms in the body has recently attracted public and media attention. Probiotics, beneficial microorganisms typically found in the gut and other organs, help digest food, absorb nutrients and produce essential vitamins such as vitamins B and K.
Additionally, they play a crucial role in regulating the immune system and reducing inflammation. This has led to the concept of smart probiotics, in which probiotics are combined with cutting-edge synthetic biological technologies such as biosensors and gene editing, representing an exciting frontier in healthcare.
The most critical aspect of smart probiotic technology is the sensors. Since biological systems function as complex networks with precise interactions, it is essential to monitor changes in the intestinal environment in real time and control microbial activity accordingly. However, challenges such as biocompatibility, precision, and sensitivity limit the types of components that can be used in these sensors, making technology development difficult.
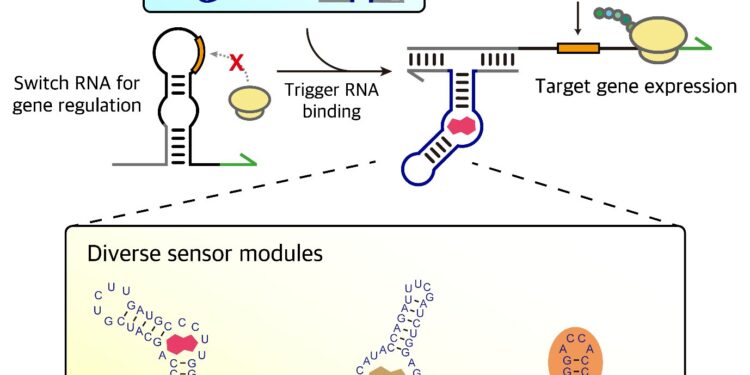
To solve this problem, Professor Jongmin Kim’s team developed the START technology platform based on aptamers. Aptamers are short strands of nucleic acids (DNA or RNA) that bind specifically and strongly to target molecules, making them ideal components for sensors requiring high selectivity.
When an aptamer binds to its target, it triggers a structural change in the sensor aptamer, such as folding or unfolding. The team exploited this mechanism to create a platform that mimics natural biological interactions, detects various molecules, and regulates gene expression as easily as assembling LEGO blocks.
Building on this foundational technology, the team successfully developed sensors capable of detecting drugs and antibiotics like theophylline and tetracycline as well as specific proteins like MS2 produced by bacteria.
These sensors are designed with both the specificity to accurately recognize and respond to their targets and the orthogonality to operate independently without interference, even when multiple sensors are used together. This enables seamless monitoring and regulation of various biological signals, including logic circuits integrated into complex genetic networks.
Compared to existing technologies, the team’s platform offers significant engineering potential. This platform allows tunable control of the sensitivity and intensity of the sensor response rather than simple detection of target biomolecules. Notably, the team was able to create new biosensor components that follow simple design rules, unlike their natural counterparts in biological systems.
Professor Jongmin Kim from POSTECH said: “This study opens new possibilities for the development of synthetic biosensors and the design of microbial genetic circuits. We will continue to explore applications in microbial engineering technologies, including smart probiotics and metabolic engineering. »
In addition to Professor Kim, researchers include Ph.D. candidates Jeongwon Kim, Minchae Seo and Yelin Lim from the Department of Life Sciences at POSTECH.
More information:
Jeongwon Kim et al, START: A versatile platform for bacterial ligand detection with programmable performance, Advanced science (2024). DOI: 10.1002/advs.202402029
Provided by Pohang University of Science and Technology
Quote: LEGO-inspired RNA sensors enable customizable genetic control (October 2, 2024) retrieved October 2, 2024 from
This document is subject to copyright. Except for fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for informational purposes only.