In situ genome sequencing (ExIGS) expansion workflow. Credit: bioRxiv (2024). DOI: 10.1101/2024.09.24.614614
Harvard scientists have unveiled a new technique called expansion in situ genome sequencing (ExIGS) that combines existing in situ genome sequencing (IGS) with expansion microscopy (ExM). The innovation allowed researchers to link abnormalities in the nucleus to alterations in gene regulation within a single cell through precise measurements of DNA-protein interactions at the nanoscale.
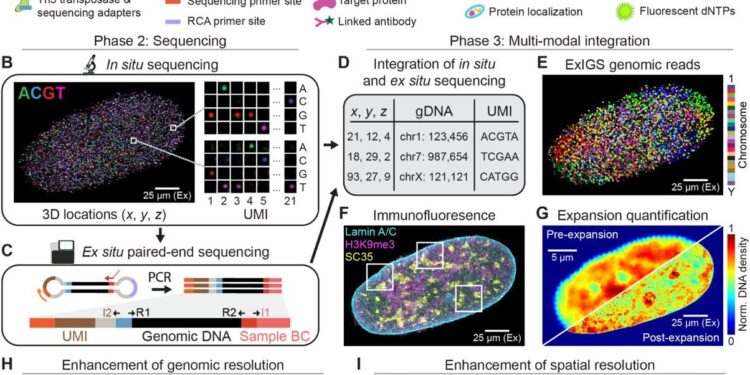
Microscopy is an essential tool for characterizing cellular function. Imaging methods are limited by the diffraction limit of optical microscopy, preventing reliable scale measurement of DNA-protein interactions.
Expansion microscopy is a clever technique that overcomes the problem of seeing objects hidden by the diffraction limit by enlarging them. By embedding the sample in a swellable polymer gel and expanding it, researchers can obtain super-resolution imaging of spatial organizations within cells using conventional microscopes.
In ExIGS, expansion microscopy is integrated with in situ genome sequencing to simultaneously sequence genomic DNA and image nuclear proteins at nanometer resolution in single cells.
The process begins by embedding fixed cells in a polyacrylate-based gel, which serves as a scaffold for expansion. The genomic DNA and proteins are chemically linked to the gel using DNA oligohooks that bind to the DNA hairpins attached to the genomic fragments and proteins. These hooks contain chemical groups that form covalent bonds with the gel during polymerization.
Once incorporated, the gel polymerizes around attached cellular components, immobilizing DNA and proteins. Water is added to the gel, causing it to swell evenly about 4.5 to 5.5 times in all directions. This expansion physically enlarges the cell while preserving the spatial relationships between molecules. To maintain the integrity of the newly expanded structure, the sample is then re-embedded in a non-expandable secondary acrylamide gel.
The expanded sample undergoes super-resolution immunofluorescence imaging, allowing detailed visualization of nuclear proteins. Rolling circle amplification is then performed to generate clonal DNA amplicons, which are sequenced in situ. This amplification ensures precise sequencing and localization of the genomic DNA in the expanded nucleus.
The researchers detail testing of their innovation in a preliminary paper titled “In situ expansion genome sequencing links nuclear abnormalities to hotspots of aberrant euchromatin repression,” on the topic. bioRxiv preprint server.
By applying ExIGS to fibroblastic cells from a patient with Hutchinson-Gilford progeria syndrome, researchers identified irregularities in nuclear lamins (proteins that structure the nucleus) that create hotspots of abnormal repression in active genomic regions , potentially compromising cellular identity.
The study demonstrated that ExIGS significantly increases the number of genomic reads per core and preserves 3D genome structure comparable to Hi-C sequencing data.
The technique provides a powerful new platform for exploring DNA-protein interactions, allowing researchers unprecedented insight into molecular mechanisms. This could significantly improve our understanding of a wide range of diseases and genetic disorders, reveal the hidden interactions of cellular senescence in aging, and become a breakthrough technology in biotechnology research laboratories.
More information:
Ajay S. Labade et al, Expansionin situgenome sequencing links nuclear abnormalities to hotspots of aberrant euchromatin repression, bioRxiv (2024). DOI: 10.1101/2024.09.24.614614
© 2024 Science X Network
Quote: Expanding in situ genome sequencing innovation makes hidden DNA-protein interactions visible (October 14, 2024) retrieved October 14, 2024 from
This document is subject to copyright. Except for fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for informational purposes only.