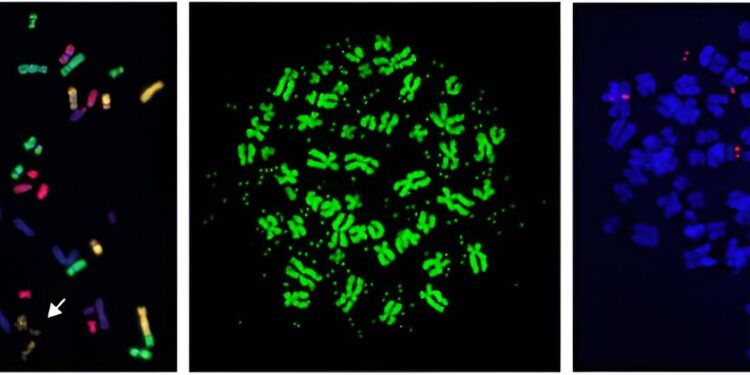
These images show examples of mitotic chromosome spread from human cancer cells containing a broken chromosome (left panel) or extrachromosomal DNA (middle and right panels), which fuel cancer genome evolution and resistance to anticancer drugs. Credit: UT Southwestern Medical Center
Cancer cells appear to hijack a genetic pathway involved in DNA repair to promote malignancy and overcome treatment, a study by researchers at UT Southwestern Medical Center shows. Their findings, published in Cellexplain how chromosomes in some tumors undergo massive rearrangements and could lead to new strategies to avoid resistance to cancer drugs.
“Our research addresses a key mechanistic question in cancer biology by identifying a source of chromothripsis, a mutational process driven by chromosome fragmentation. Chromothripsis allows cancer cells to rapidly evolve their genome by dramatically rearranging individual chromosomes, leading to multiple genetic alterations in a relatively short period of time,” said study leader Peter Ly, Ph.D.
Ly is an assistant professor of pathology and cell biology at UT Southwestern and a member of the Cellular Networks in Cancer Research Program at the Harold C. Simmons Comprehensive Cancer Center.
Chromosomal rearrangements caused by chromothripsis occur in 30–40% of all cancers and are typically seen in aggressive tumors such as sarcomas, glioblastomas, and pancreatic cancer.
Chromothripsis can occur when mitosis, the process of cell division, goes wrong. Cells can mistakenly sort entire chromosomes or arms of chromosomes out of the nucleus and into abnormal bundles called micronuclei. Previous work from the Ly Lab has shown that these chromosomes in micronuclei eventually break into multiple pieces and reassemble in the wrong order. How these chromosomes break apart has not been identified.
To answer this question, Dr. Ly, in collaboration with Justin Engel, BS, a graduate student researcher in the Cancer Biology PhD program, used CRISPR, a gene-editing tool, to inactivate genes in cells with micronuclei to find those that might play a key role in chromosome destruction.
Their research focused on a set of genes involved in the Fanconi anemia pathway, a DNA repair mechanism mutated in an eponymous germline disease characterized by severe anemia, bone marrow failure, a predisposition to cancer and other birth defects. When the researchers inactivated these genes, the chromosomes did not break.
Further research showed that, unlike the chromosomes in the nucleus, those in the micronuclei fail to replicate properly, triggering the Fanconi anemia pathway. As part of this DNA repair process, an enzyme complex then cuts these chromosomes into pieces, causing the chromosome fragmentation characteristic of chromothripsis.
“When these fragments are stitched together in the wrong order,” Dr. Ly explains, “they cause chromosomal rearrangements that often disable genes that are supposed to protect against cancer development. In addition, some fragments develop into a circular DNA structure called extrachromosomal DNA, or ecDNA, which is amplified into large numbers of copies, a process that can promote drug resistance.”
To extend their findings to a relevant clinical setting, Dr. Ly and colleagues inactivated the Fanconi anemia pathway in melanoma tumor cells treated with targeted therapy. Although these cancer cells typically acquire resistance to these drugs over time, cells lacking the Fanconi anemia pathway did not undergo chromothripsis and did not develop drug resistance.
“These findings could potentially lead to new strategies incorporating Fanconi anemia pathway inhibition with other drugs to combat the emergence of treatment-resistant cancer cells, which is a significant problem for cancer patients,” Dr. Ly said.
More information:
Justin L. Engel et al, The Fanconi anemia pathway induces chromothripsis and ecDNA-mediated cancer drug resistance, Cell (2024). DOI: 10.1016/j.cell.2024.08.001
Cell
Provided by UT Southwestern Medical Center
Quote:A pathway linked to genomic alterations responsible for cancer has been identified (2024, September 10) retrieved September 10, 2024 from
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without written permission. The content is provided for informational purposes only.