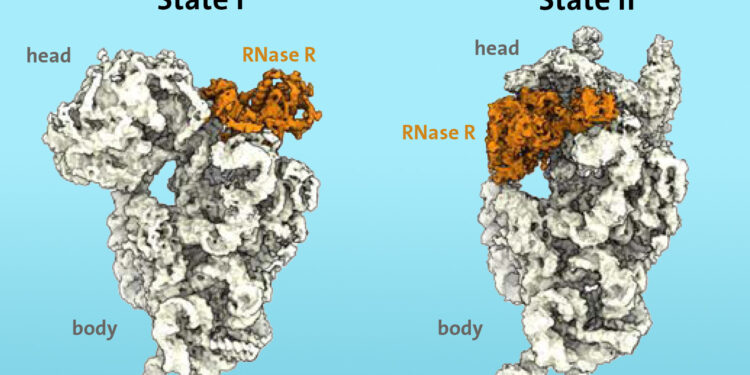
Degradation of the 30S subunit of the ribosome. Credit: UHH/MIN/Paternoga
A research team from the Department of Chemistry at the University of Hamburg has succeeded for the first time in identifying at the molecular level the dynamic mechanism used by the RNase R enzyme to degrade the 30S ribosomal subunit. The results of the study were published in the scientific journal Nature.
Protein synthesis is a vital and energy-intensive process in the cell in which ribosomes play a crucial role. These relatively large molecules are found in all living organisms and act as the “protein factories” of cells.
To do this, ribosomes read the pattern of a specific protein on a messenger molecule – messenger RNA (mRNA) – then convert this information into a new protein. Ribosomes are made up of two subunits. The small subunit is responsible for reading and checking for errors in the mRNA, while the large subunit is responsible for polymerizing amino acids to form proteins.
Controlled production and regulated turnover of ribosomes are necessary for protein synthesis. Although ribosome assembly has become increasingly understood in recent years, there is no structural insight into ribosome degradation.
This is important because in stressful situations like lack of food or at the end of their growth cycle, cells reduce their metabolism in order to survive longer. This state is called stationary phase. During this phase, energy-intensive protein synthesis is reduced, and certain ribosomes are degraded in order to release the energy invested in them to ensure cell survival.
For their investigations, the researchers studied Bacillus subtilis, a rod-shaped soil bacterium found in air, dust and water as well as in the intestines of humans and animals. “Unlike previous studies, we took cells that were still growing and were not in the stationary phase. We wanted to know what processes take place during the transition to the stationary phase,” explains Dr. Helge Paternoga from the Department of chemistry from the University of Vienna. Universität Hamburg, last author of the study.
Researchers knew from previous work that certain enzymes, such as ribonuclease R (RNase R), are involved in the process of ribosome degradation under stress. Using cryo-electron microscopy, they were able to show for the first time that the RNase R enzyme binds to the small 30S subunit of the ribosome. The “S” stands for “Svedberg units” and refers to the mass of the ribosomal subunit.
RNase R does not arbitrarily cut the 30S subunit, but rather attaches to a free area, which researchers call the “neck,” then detaches the “head,” the top area of the subunit, in two consecutive steps.
“In the first step, the RNase R enzyme encounters an obstacle at the ‘neck’ and destabilizes the neck area, making it more flexible. In the second step, the ‘head’ is rotated, removing the obstacle and allows the enzyme to continue the process of degrading the 30S subunit unhindered,” explains Paternoga.
“Our in vitro degradation experiments indicate that the head switch constitutes an important kinetic barrier for RNase R. Furthermore, we were able to show that the enzyme alone is sufficient to accomplish the complete 30S degradation process,” explains Professor Daniel. Wilson, head of the research group at the Department of Chemistry at the Universität Hamburg and co-author of the study.
More information:
Lyudmila Dimitrova-Paternoga et al, Structural basis of degradation of the 30S ribosomal subunit by RNase R, Nature (2024). DOI: 10.1038/s41586-024-07027-6
Provided by the University of Hamburg
Quote: First molecular insights into the degradation of the 30S ribosomal subunit (February 8, 2024) retrieved February 8, 2024 from
This document is subject to copyright. Apart from fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.