Overview of study design and analytical approaches. Credit: Natural medicine (2024). DOI: 10.1038/s41591-024-03164-7
Researchers have developed a machine-learning-based blood test that analyzes more than 200 proteins to assess a person’s rate of biological aging, which the team says can be used to estimate a person’s risk of developing 18 major age-related diseases and dying prematurely from any cause.
This work helps validate the use of the proteome (all the proteins present in the body at a given time) as a precise indicator of a person’s age, not in years, but in terms of the functioning of their cells.
These findings provide insight into the biological pathways that lead a person to develop multiple age-related diseases, pave the way for a better understanding of how genes and the environment interact in aging, and could help researchers develop treatments for age-related diseases and evaluate their effectiveness.
Although the test is currently limited to the research lab, the team is working to develop it into something anyone can order in a doctor’s office.
Austin Argentieri, a medical researcher in the Analytical and Translational Genetics Unit at Massachusetts General Hospital, is the lead author of the study, published Aug. 8 in Natural medicine and discusses his team’s findings below.
What question did you seek to answer with this study?
Can we develop a proteomic aging clock that could help predict the risk of common age-related diseases?
Age is the primary determinant of most common chronic diseases, but it is an imperfect proxy for aging, which is the driver of age-related multimorbidity (having more than one chronic health condition) and mortality.
Aging can be more accurately estimated by using “omics” data to capture an individual’s biological functioning relative to an expected level of functioning for a given chronological age.
While the most common biological clocks of aging use DNA methylation, protein levels can provide more direct mechanistic and functional insight into the biology of aging. Moreover, the proteome is the most common target for drug development.
However, previous studies of the proteomic aging clock have not been independently validated among populations with diverse genetic and geographic backgrounds.
To date, no test has been developed in large or well-powered general population samples that would allow association testing across a broad spectrum of age-related disorders, multimorbidity, and mortality.
What did you find?
We developed a machine learning model that uses blood proteomic information to estimate a proteomic age clock in a large sample of participants from the UK Biobank. Our sample included 45,441 participants aged 40–70 years.
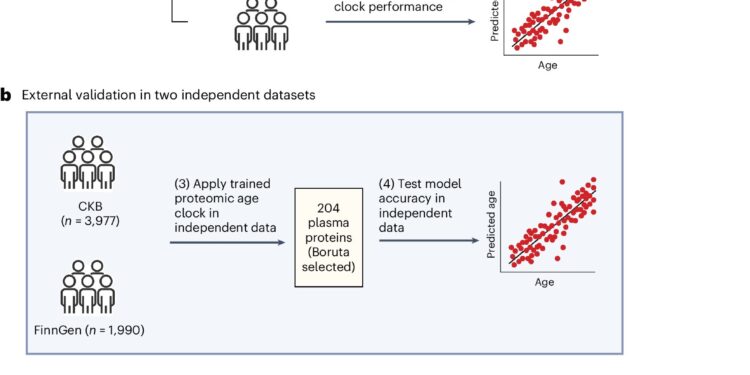
We then validated this model in two biobanks around the world: 3,977 participants aged 30–80 years from the Chinese Kadoorie Biobank and 1,990 participants aged 20–80 years from the FinnGen Biobank in Finland. These biobanks are geographically and genetically distinct populations that have different age ranges and disease profiles than the UK biobank.
We identified 204 proteins that accurately predict chronological age, and we further identified a set of 20 aging-related proteins that capture 91% of the age prediction accuracy of the larger model.
We demonstrated that our proteomic age clock exhibited similar age prediction accuracy in independent participants from China and Finland compared to its performance in the UK Biobank.
We found that proteomic aging was associated with the incidence of 18 major chronic diseases, including heart, liver, kidney and lung diseases, diabetes, neurodegeneration such as Alzheimer’s disease, and cancer, as well as multimorbidity and all-cause mortality risk.
Proteomic aging was also associated with age-related measures of biological, physical, and cognitive function, including telomere length, frailty index, and several cognitive tests.
What are the clinical implications of your work?
We provide some of the largest and most comprehensive evidence to date demonstrating that proteomic aging is a common biological signature linked to many age-related functional traits, morbidities, and mortalities.
We also provide some of the first evidence that a proteomic age clock may be highly generalizable across human populations of diverse genetic ancestries, age groups, and disease profiles.
Multimorbidity is a major problem in clinical and population health that has a major impact on health care costs. Our proteomic clock gives us a first insight into the pathways that form the biological basis of multimorbidity.
In the near future, proteomic aging clocks can be used to study the relationship between genetics and environment in aging, providing new insights into the drivers of aging and multimorbidity across the lifespan.
An important avenue will also be to use proteomic clocks as a biomarker of the effectiveness of preventive interventions targeting aging and multimorbidity.
Additionally, proteomic clocks can be used to accelerate drug development and clinical trials by identifying high- and low-risk patients. For example, less than 1% of people in the bottom decile of proteomic aging developed Alzheimer’s disease within 10 to 15 years.
More information:
M. Austin Argentieri et al, Proteomic aging clock predicts mortality and risk of common age-related diseases in diverse populations, Natural medicine (2024). DOI: 10.1038/s41591-024-03164-7
Provided by Harvard Medical School
Quote:Experimental blood test predicts age-related disease risk in diverse populations (2024, August 16) retrieved August 16, 2024, from
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without written permission. The content is provided for informational purposes only.