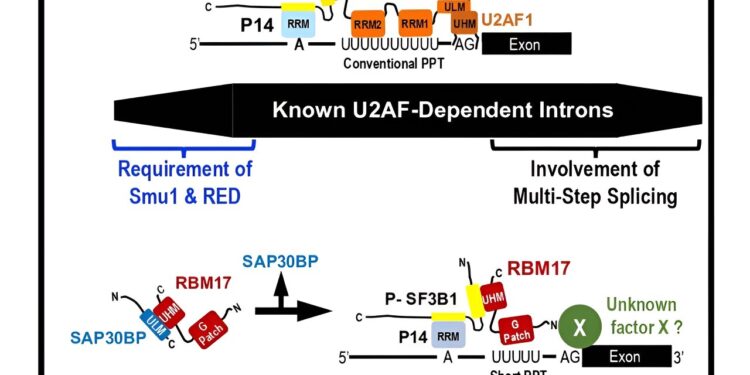
Human introns vary greatly in length. Previously, only U2AF-dependent splicing was known. Researchers at Fujita Health University now show that splicing in a subset of human short introns is mediated by the RBM17–SAP30BP intermediate complex, instead of the U2AF heterodimer. Credit: Kazuhiro Fukumura and Akila Mayeda of Fujita Health University, Japan. Cell Reports (2023). DOI: 10.1016/j.celrep.2023.113534
The well-known essential heterodimer of pre-mRNA splicing factor U2AF (U2AF2–U2AF1) was identified to mediate early splicing reactions in all introns of different lengths. However, Dr. Kazuhiro Fukumura from the Akila Mayeda laboratory at Fujita Health University discovered that a subset of short introns with truncated polypyrimidine tracts are spliced by the RBM17–SAP30BP complex instead of the U2AF heterodimer. Dr. Fukumura’s team proposed a unique mechanism in which SAP30BP guides RBM17 to active early spliceosomes.
In humans, the length of the pre-mRNA varies considerably (from 30 to 1,160,411 nucleotides according to recent studies). The fundamental mechanism of splicing was studied with pre-mRNA models including for example 158 and 231 nt introns, which are spliced very efficiently in vitro and in vivo.
Such an ideal pre-mRNA contains good splicing signal sequences, i.e., 5′ splice site, branch site (BS) sequence and polypyrimidine tract (PPT) followed by site 3′ splicing which are recognized by the U1 snRNP. U2 snRNP and U2AF2–U2AF1, respectively. Professor Mayeda says: “Given the varying lengths of human introns, it is likely that there is more than one mechanism. This is our motivation to initiate our study of splicing focused on human short introns.
Dr. Fukumura explains: “Our previous research on the splicing process on short introns revealed that the authentic splicing factor U2AF2 cannot bind to the truncated PPT and that RBM17 is then replaced by U2AF to promote splicing. . You know, this is reasonable because short introns are often too tight for the sufficient duration of the PPT. We published this result in 2021.
“However, RBM17 cannot bind the truncated PPT in vitro, so we did not know how the truncated PPT and the following 3′ splice site are recognized by RRM17. Therefore, we hypothesized that a Another protein factor is involved in RBM17-dependent splicing.”
The Mayeda group finally identified this protein cofactor behind RBM17-dependent splicing, which is SAP30BP. Their study was published in the journal Cell Reports on December 7, 2023.
Dr. Fukumura says: “It was essential to study previous references. From three papers, I was convinced that SAP30BP is the strongest candidate for the cofactor of RBM17. » They showed that the existence of SAP30BP in the human early splicing complex, fruit fly SAP30BP and RBM17 were detected in a fly spliceosome formed on a short intron, and that the binding between SAP30BP and RBM17 was in effect was detected by yeast two-hybrid analysis.
“Nowadays, siRNA-induced depletion of SAPBP in human cell line is the easiest and most straightforward way to verify RBM17-dependent splicing repression. And it was bingo!” Dr. Fukumura said.
Transcripts in SAP30BP-depleted human cells were analyzed by a next-generation sequencer (RNA-Seq analysis), and many RBM17- and SAP30BP-dependent introns were found. These introns were distributed over a shorter range and the truncated PPT was indeed a critical determinant of RBM17/SAP30BP dependence. Thus, RBM17 and SAP30BP are the general splicing factors.
Professor Mayeda remarks: “It was a happy coincidence that Professor Michael Sattler, an expert in structural analyses, was keenly interested in our study, and we were able to begin a productive collaboration.”
Protein-protein interactions via UHM (U2AF homology motif)–ULM (UHM motif-ligand) binding play an essential role in general splicing reactions. The Sattler lab discovered a hidden UHM sequence in SAP30BP and demonstrated that it was essential for interacting with the UHM in RBM17 by NMR (nuclear magnetic resonance) and ITC (isothermal titration calorimetry) analyses.
However, the role of the RBM17–SAP30BP interaction remains enigmatic. Since RBM17 has only one UHM, the RBM17–SAP30BP bond must be released before the interaction of RBM17 with SF3B1, a component of the U2 snRNP, essential to promote splicing. So, what is the role of the RBM17-SAP30BP interaction?
Professor Mayeda says: “Fukumura designed a clever binding assay using anti-phospho-SF3B1 antibodies to answer this curious question, and we could provide an elegant working model. »
The investigators propose that the RBM17–SAP30BP intermediate complex prevents nonfunctional binding of RBM17 to free, unphosphorylated SF3B, which promotes functional binding of RBM17 to active phosphorylated SF3B1 on the pre-mRNA.
More information:
Kazuhiro Fukumura et al, SAP30BP interacts with RBM17/SPF45 to promote splicing in a subset of human short introns, Cell Reports (2023). DOI: 10.1016/j.celrep.2023.113534
Provided by Fujita Health University
Quote: Discovery of a subset of human short introns separated by a new mechanism (February 14, 2024) retrieved February 14, 2024 from
This document is subject to copyright. Apart from fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.