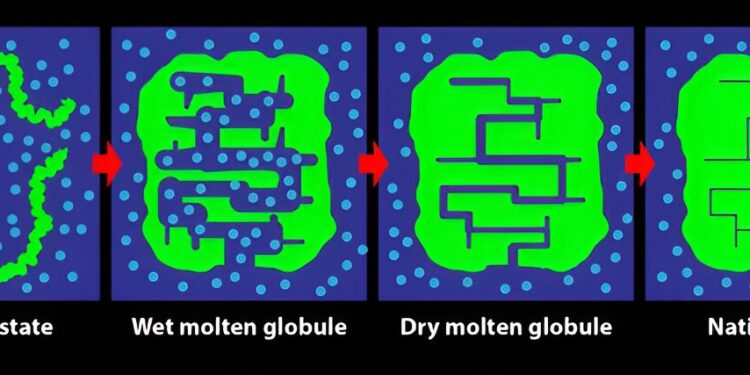
A protein in four different states when it folds. (The blue circles are water molecules.) The dry molten globule state, recently observed in this study, has water extruded from the interior of the protein but is still not in its final folded state , which is the rightmost configuration in the figure. Credit: APS/Alan Stonebraker
Scientists have discovered a new intermediate state in the protein folding process, showing that folding can occur in two steps, one fast and the next much slower. The results are published in the journal Physical Examination Letters.
After a protein – a string of up to 20 different amino acid molecules – is created in a cell, it spontaneously coils (“folds”) from a random structure into an ordered three-dimensional structure that makes it biologically useful. (Incorrect folding can lead to diseases such as amyotrophic lateral sclerosis, ALS, also known as Lou Gehrig’s disease.)
For simpler proteins, this usually happens very quickly, in about 0.01 milliseconds (ms), and takes its almost complete form. In this research, a second, slower state, lasting 3 to 10 milliseconds, occurred after the first step, up to a thousand times longer than the previous folding, due to small rearrangements of the side chains on the protein. (In contrast, the blink of a human eye lasts between 100 and 400 milliseconds.)
Scientists have learned to predict, using machine learning, the configuration a protein will fold into. But it is poorly understood why each particular structure, different for each protein, is obtained, and how this happens when different amino acids react chemically with each other.
Previous analytical methods have, through methods such as optical spectroscopy (which measures the difference between how left- and right-handed circularly polarized light is absorbed), examined how proteins extract water from inside as they turn and twist into their coiled and folded form. State. However, such optical measurements do not capture the final long-term folding discovered here.
Researchers at the US National Institute of Health’s National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), led by Robert Tycko, acting head of its Chemical Physics Laboratory, decided to research such declines in the longer term.
They chose the HP35 protein, known for its simplicity and rapid folding, consisting of 35 amino acid residues. (Residues are the result of a bond between two amino acid molecules, forming a peptide, which removes water. These residues influence other residues and the resulting chemical properties of the entire protein.) Also known as Named “nasty helmet subdomain”, it is a protein commonly used in folding studies.
The group used heat to unfold the HP35, heating it for 25 ms to a warm temperature of 95°C. They then allowed it to fold for an “incubation period” of about 0.1 ms at a warm temperature of 30°C. They held the protein at this temperature for an incubation period of 1 to 10 milliseconds, then froze it at -35°C, where particles approximately 0.3 millimeters in diameter formed.
After observing the rapid folding of HP35 using optical spectroscopic probes, the frozen particle suspension was analyzed by solid-state nuclear magnetic resonance of carbon-13 atoms in the protein residues to observe their behavior after folding quick initial. HAS 13Oscillation frequencies characteristic of the nuclear spin of C, the oscillating applied magnetic field signal becomes sharper as the molecular structure becomes more rigid.
The group observed that the 13The C resonance frequencies for two specific residues did not change during the incubation period, indicating that these were sections of the protein that had already folded at the start of the incubation period. However, the signal from the resonance frequencies of several other residues in other parts of the protein grew sharper over the 10 ms incubation period, revealing that these protein sections were still folding after the start of the incubation period.
That is, HP35 appeared bent over 0.1 ms at the start of the incubation period, but actually only became fully bent after the end of the 10 ms incubation period, becoming rigid in its final form.
This result is the first definitive evidence for the formation of the so-called DMG (dry molten globule) state during folding. DMG was first proposed theoretically as the penultimate state taken by a folding protein in 1989. Until then, only the wet molten globule state, with the protein preformed in a larger configuration in water, had been observed.
Until now, there was no evidence for DMG, but some suggested it existed in studies of the unfolding of various proteins. Another study of an enzyme – catalytic proteins that speed up chemical reactions – provided clues that folding could continue over several days.
“The second step of structural optimization or annealing has not been observed in previous studies but appears in our experiments because we measure NMR signals,” said Tycko of HIDDK, “which are very sensitive to conformational variations and to local structural details.
Noting that much of the equipment to measure sub-millisecond temperature jumps was developed in his lab, he added that “with this equipment we can look for similar effects in other biomolecular processes involving significant unidirectional structural changes.”
More information:
C. Blake Wilson et al, Experimental evidence for structural evolution in milliseconds – time scale after folding in microseconds – time scale of a small protein, Physical Examination Letters (2024). DOI: 10.1103/PhysRevLett.132.048402
© 2024 Science X Network
Quote: Detection of a new state in the protein folding process (February 15, 2024) retrieved on February 15, 2024 from
This document is subject to copyright. Apart from fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.