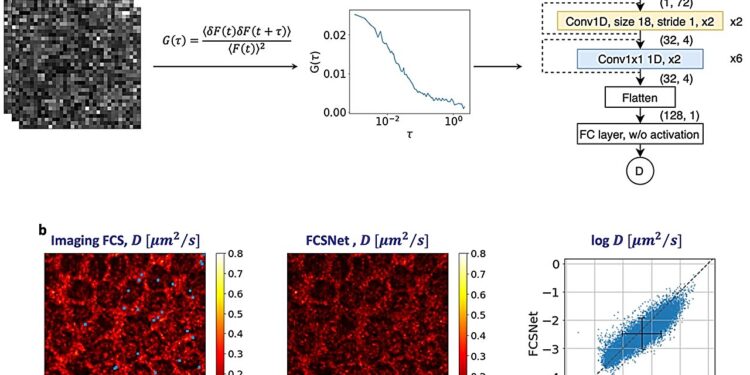
(a) FCSNet is a convolutional neural network (CNN), which takes into account the autocorrelation function of a measurement as input to predict the diffusion coefficient. (b) FCSNet enables evaluations on complex samples with as good or better accuracy than traditional FCS imaging methods, as shown by heatmaps. Credit: Biophysical Journal
Researchers at the National University of Singapore (NUS) have demonstrated that deep learning allows them to observe the dynamics of single molecules more precisely and with less data than traditional assessment methods. They used convolutional neural networks (CNNs) to observe the movement of single molecules in artificial systems, cells and small organisms. Their findings were published in the Biophysical Journal.
This method promises to speed up measurements of single molecules in complex systems and make it more accessible to a wider range of researchers.
A single molecule is the most fundamental unit observable in biological systems. Understanding its behavior and interactions provides a better understanding of how biological systems work, paving the way for strategic interventions against diseases.
One of the most powerful ways to observe single molecules is through fluorescence spectroscopy. This is due to its strong signal and specificity, which allows only labeled molecules to be observed.
For over 50 years, fluorescence correlation spectroscopy (FCS) has been used in this field, measuring the mobility and interaction of molecules with high accuracy and precision. A recent extension of this technique, Imaging FCS, extends its capabilities to characterize mobilities, concentrations and interactions among other parameters in entire images.
Despite its capabilities, Imaging FCS poses challenges because it requires large amounts of data (approximately 100 MB for each measurement). This requires extensive computer processing, resulting in slow evaluations.
A research team, led by Professor Thorsten Wohland from the Department of Biological Sciences and the Department of Chemistry, and Professor Adrian Röllin, from the Department of Statistics and Data Science, both at NUS, used learning methods in depth to reduce the amount of data. required for a measurement (approximately 5 MB per measurement) while obtaining results comparable to traditional methods.
The technique uses two CNNs, called FCSNet and ImFCSNet developed by research team members Dr. Wai Hon Tang and Mr. Shao Ren Sim. CNNs represent a type of deep learning algorithm suitable for analyzing visual data. They use several layers of specialized filters that scan the image for specific features, such as edges, textures and colors.
By gradually extracting and combining these features, they gain a better understanding of the image, allowing them to recognize patterns and objects in visual data.
Methods based on FCSNet and ImFCSNet are more accurate than traditional FCS methods in terms of diffusion coefficients, but they have different tradeoffs in terms of data requirements and spatial resolution. By using less data, these techniques can potentially reduce evaluation time by orders of magnitude, especially for large datasets or complex systems.
Professor Wohland said: “These CNNs are trained on simulated data and can predict diffusion coefficients from much smaller datasets compared to traditional FCS methods. They are also model independent and can be used with any microscope setup. »
Dr Tang added: “CNNs are revolutionizing data analysis, providing accelerated and simplified evaluation processes. While it is important to validate their performance, CNNs have the potential to make powerful techniques available to the broader research community.
The team hopes their technique can open up new possibilities for accelerating single-molecule research and making the technology accessible to a wider range of users. CNNs do not require expert input and are poised to democratize the FCS.
More information:
Wai Hoh Tang et al, Deep learning reduces data requirements and enables real-time measurements in FCS imaging, Biophysical Journal (2023). DOI: 10.1016/j.bpj.2023.11.3403
Provided by the National University of Singapore
Quote: Deep learning for real-time molecular imaging (February 8, 2024) retrieved February 8, 2024 from
This document is subject to copyright. Apart from fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.