A quantitative gain-of-function assay for synaptic partner matching. Credit: Cell (2024). DOI: 10.1016/j.cell.2024.06.022
Hundreds of millions of years ago, single cells joined together to form multicellular organisms. At the base of this multicellular world is the cell surface: the plasma membrane that surrounds each cell, where individual units meet and communicate with each other using a language of molecules and proteins.
For biologists, being able to decipher the cellular conversation that takes place on the surface—which proteins are present, how they interact, and what changes they undergo—is key to understanding how an organism functions and, ultimately, how disease occurs.
Now, researchers at HHMI’s Janelia Research Campus, Stanford University, and the Broad Institute are developing user-friendly tools to help scientists translate this cellular communication and uncover new details about how cells exchange information.
“If you think of the body as a society of cells, what we really want to do is try to decode the language of that society and how cells communicate with each other,” says Jiefu Li, head of the Janelia group and lead author of three new papers. “We want to develop methods to understand that language and use them, step by step, to decode it.”
This new research not only gives scientists new insights into cellular communication, but also highlights the potential of proteomics (the study of all proteins in living cells and organisms) to read the basic linguistic units of the cell and unravel some of biology’s greatest mysteries.
“We started by creating tools for proteomics, and then used them to answer some biological questions to make discoveries and demonstrate the power of proteomics,” Li says. “We are demonstrating that proteomics can be applied to many biological questions: from neuroscience to immunology, and from flies to humans.”
Learning the language of cells
Li began developing methods and tools to study proteins on the surface of cells while he was a graduate student and postdoctoral fellow at Stanford. Biologists around the world began using these methods, but many of these researchers contacted Li and his colleagues with the same problem: Without training in chemical biology or proteomics, they were struggling to make sense of the data they were generating.
When Li joined Janelia as group leader in 2022, he made it his mission to solve the problem. He collaborated with the scientific computing software team to develop PEELing, a user-friendly platform that allows researchers around the world to analyze spatial proteomics data with the click of a mouse.
Li says the development of PEELing would only have been possible at Janelia, where biologists can easily collaborate with computer scientists to develop tools that benefit the entire scientific community.
“The primary goal is to help people who don’t have a background in chemical biology or proteomics use our method and to welcome people into this methodology,” Li says. “We need to make this first step simple and robust, because people want to focus on their biological discoveries instead of dealing with all the technical details.”
Deciphering the signals
Li and his colleagues are also using some of these same methods and tools to learn more about how communication occurs inside and outside the cell surface.
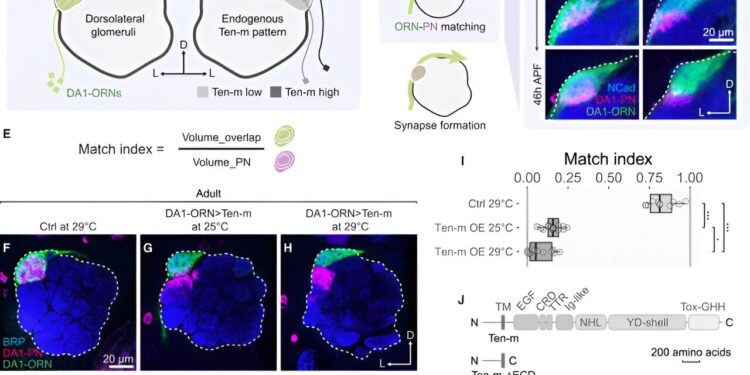
In a recent article published in CellLi and researchers at Stanford and the Broad Institute studied the signaling that occurs inside neurons after the activation of tientin, a protein that spans the cell membrane and connects axons to dendrites to form synapses in the developing brain. The scientists knew that tientin was responsible for this synaptic match, but they didn’t know what happened inside the cell after the corresponding signal was released.
The researchers first used proximity tagging to capture molecules interacting with tenurin inside the cell. They then used proteomics to identify these proteins and determine which ones were potentially involved in the process. The team then used genetics to understand how these candidate proteins interacted with tenurin, allowing the researchers to identify signaling partners and elucidate the cellular and molecular mechanisms behind the synaptic partner match.
In another recent study published in Journal of immunologyLi and researchers from Stanford and the Broad Institute used cell surface proteomics to examine human dendritic cells, which coordinate the innate and adaptive immune systems through their cell surface proteins. To better understand this coordination, the team studied how the cell surface changes when dendritic cell activation occurs.
The researchers activated the dendritic cells, then labeled, captured, and quantified proteins on the cell surface. They then annotated these proteins according to their known functions and compared the proteins present in activated and resting dendritic cells. Through this characterization, the researchers discovered how the cell modulates the activity of dozens of proteins to regulate the immune response.
“Overall, all of this work focuses on our main goal in the lab, which is to decode the cell surface,” Li says. “Proteomics can provide a comprehensive systems overview and very informative insights for follow-up mechanistic studies.”
More information:
Chuanyun Xu et al, Molecular and cellular mechanisms of tientin signaling in synaptic partner matching, Cell (2024). DOI: 10.1016/j.cell.2024.06.022
Namrata D. Udeshi et al, Cell surface milieu remodeling in human dendritic cell activation, Journal of immunology (2024). DOI: 10.4049/jimmunol.2400089
Provided by the Howard Hughes Medical Institute
Quote:Decoding the language of cells using the power of proteomics (2024, September 5) retrieved September 5, 2024 from
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without written permission. The content is provided for informational purposes only.