CausalXtract Pipeline. Credit: eLife (2024). DOI:10.7554/eLife.95485.1
Researchers have developed a tool that provides new insights into the cause-and-effect relationships between cells and how they change over time.
The research, published today as a revised preprint in eLifeis described by the editors as a seminal study introducing a new data processing pipeline that could be used to better understand cell-cell interactions. The utility of this pipeline is convincingly illustrated using tumor ecosystem-on-a-chip data, but it could also be applied to perform causal discovery in other scientific fields, meaning this work could potentially have a wide range of applications.
The ability to image living cells under different experimental conditions has made it possible to extract valuable information about the shape and state of cells and their interactions with other cells. But this wealth of information remains underexploited because, until now, there has been a lack of methods and tools to identify cause-and-effect relationships between observed features. This ability to identify cause-and-effect relationships is called causal discovery.
The new tool, called CausalXtract, was adapted from a previous discovery method that can learn causal networks of biological systems but without information about the timing of events.
“Our previous causal discovery tool can learn contemporary causal networks for a wide range of biological or biomedical data, from single-cell gene expression data to patient medical records,” says co-lead author Franck Simon, a research engineer at Institut Curie, Université PSL, Sorbonne Université, France.
“However, time-lapse imaging data from live cells contain information about cellular dynamics, which can facilitate the discovery of new cause-and-effect processes, based on the assumption that future events cannot cause past events.”
Simon was co-lead author of the paper alongside Maria Colomba Comes, then a PhD student in the Department of Electronic Engineering at the University of Rome Tor Vergata, Rome, Italy, now a researcher at the Tumor Institute of Bari, Italy, and Tiziana Tocci, a PhD student at the Institut Curie, PSL University, Sorbonne University.
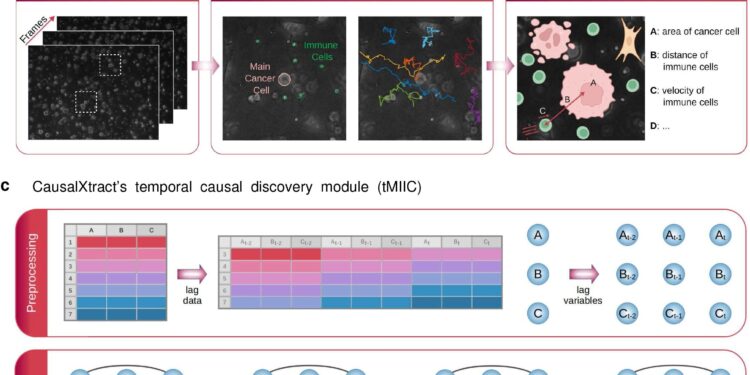
To study this question, CausalXtract reconstructs time-unfolded causal networks, where each variable is represented by multiple nodes at different times. This takes into account the links between successive time steps in the data. This graph-based causality goes beyond early models of temporal causality (“Granger causality”) that can neglect real causal effects, as the study shows.
“We evaluated the tool using artificial datasets that resemble real-world data in terms of number of time steps and network size and found that it matches or outperforms existing methods while running orders of magnitude faster,” Simon adds.
To test the tool’s performance with real biological data, the team used time-lapse image data from a tumor-on-a-chip model demonstrating the effects of the cancer drug trastuzumab. A tumor-on-a-chip model replicates the 3D structure and microenvironment of a tumor, consisting of tumor cells, immune cells, tumor-associated fibroblasts, and endothelial cells.
From this model, “we extracted raw images of cellular characteristics such as geometry, speed, cell division, cell death, and transient and persistent cellular interactions,” Tocci explains. The team then reconstructed a causal network that developed over time from information about cellular characteristics, interactions, and therapeutic conditions at different times.
“This reconstruction revealed new biologically relevant information and confirmed existing known relationships between cells,” says study co-author Maria Carla Parrini, who supervised the tumor-on-a-chip experiments at the Institut Curie.
For example, the model confirmed that trastuzumab treatment increases cell death and the number of interactions between cancer cells and immune cells, but it also showed for the first time that cancer-associated fibroblasts (CAFs) independently block cancer cell death. While CAFs have previously been reported to reduce treatment effectiveness, these results provide new insights into how this happens.
The team also noted with interest that CausalXtract identifies opposing effects at different times. For example, it was able to see that a cell’s eccentricity (the extent to which the cell deviates from its normal circular shape) changes at different stages of cell division.
Late phases of cell division are associated with an increase in cell eccentricity, but this is preceded by a decrease in eccentricity 2–4 hours before cell division, once the decision to divide has been made. This demonstrates the potential of the tool to uncover new, possibly time-lagged, causal relationships between cellular features.
“CausalXtract opens new avenues to analyze live cell imaging data for a range of basic and translational research applications, such as using tumors-on-chips to screen immunotherapy responses on tumor samples from patients,” says co-senior author Eugenio Martinelli, full professor in the Department of Electronic Engineering at the University of Rome Tor Vergata.
“With the advent of virtually unlimited live cell image data, flexible interpretation methods are essential, and we believe that CausalXtract can bring unique insights based on causal discovery to interpret these information-rich data,” adds co-lead author Hervé Isambert, group leader DR CNRS, Institut Curie, Université PSL, Sorbonne Université.
More information:
Franck Simon et al, CausalXtract: a flexible pipeline to extract causal effects from live cell time-lapse imaging data, eLife (2024). DOI: 10.7554/eLife.95485.1
Journal information:
eLife
Quote:Computer tool can identify causal relationships from complex biological data (2024, September 17) retrieved September 17, 2024 from
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without written permission. The content is provided for informational purposes only.