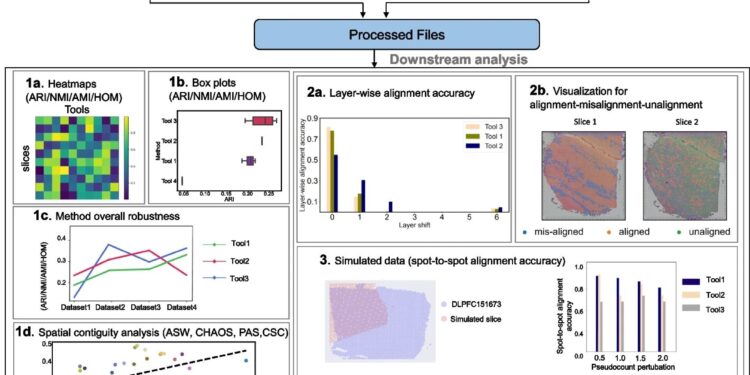
Framework for benchmarking clustering, alignment and integration methods on different real and simulated datasets. Credit: Genome biology (2024). DOI: 10.1186/s13059-024-03361-0
A team of Vanderbilt researchers has published a new comparative study that aims to help scientists select the most effective methods for analyzing spatial transcriptomics (ST) data.
The study led by Xin Maizie Zhou, assistant professor of biomedical engineering and computer science, evaluates computational tools for spatial transcriptomics (ST), a technology used to map gene expression patterns in tissues while preserving spatial context . It was recently published in Genome biology.
ST involves cutting a tissue sample and placing it on a specially designed slide with spatially indexed barcodes. When the tissue is processed, the ribonucleic acid (RNA) present at each specific location in the tissue is captured by these barcodes. After RNA sequencing, the data can be mapped to the original tissue locations, allowing researchers to visualize where certain genes are expressed in the tissue architecture.
Since its widespread use began in 2020, this revolutionary sequencing technology has revolutionized the understanding of complex tissues. Applications of ST include cancer research and neuroscience, such as mapping gene expression in parts of the brain to understand regional functions or disease mechanisms.
However, the variety of tools available for analyzing ST data can be overwhelming, making it difficult to choose the right approach for specific research needs.
To address this issue, the Vanderbilt team systematically compared 16 clustering methods, five alignment methods, and five embedding methods on a variety of data sets. Their results offer practical recommendations for researchers working with spatial transcriptomics, helping them identify the tools that best fit their research needs.
“Our goal was to provide a clear and accessible guide for researchers navigating the options available for spatial transcriptomic analysis,” said Zhou, who is on the faculty of Vanderbilt’s Trans-Institutional Data Science Institute . “We hope this study will be a useful resource for anyone working in this rapidly growing field.”
More information:
Yunfei Hu et al, Comparative analysis of clustering, alignment and integration methods for spatial transcriptomics, Genome biology (2024). DOI: 10.1186/s13059-024-03361-0
Provided by Vanderbilt University
Quote: A comparative study aims to help scientists analyze spatial transcriptomics data (October 10, 2024) retrieved on October 10, 2024 from
This document is subject to copyright. Except for fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for informational purposes only.