Graphic summary. Credit: Cell (2024). DOI: 10.1016/j.cell.2024.08.039
Scientists have identified thousands of genetic changes in a gene that can increase the risk of developing breast or ovarian cancer, paving the way for better risk assessment and more personalized care.
Researchers at the Wellcome Sanger Institute and their collaborators focused on the RAD51C gene, a “cancer protection” gene. They found more than 3,000 harmful genetic changes that could disrupt its function and increase the risk of ovarian cancer by six-fold and the risk of aggressive breast cancer subtypes by four-fold. These findings were confirmed by analysis of data from large-scale health databases.
The results, published today (September 18) in Cellare available free of charge so they can be immediately used to help doctors and scientists in diagnostic laboratories better assess cancer risk, particularly for people with a family history of these cancers, thereby reducing the uncertainty that often accompanies genetic testing.
The study also identified regions of the protein essential for its function, indicating new roles in cancer development and potential therapeutic targets.
Breast cancer is the most common cancer in the UK, with around 56,800 new cases each year. One in seven British women will be diagnosed with breast cancer in their lifetime. Ovarian cancer is the sixth most common cancer in women in the UK, with around 7,500 new cases each year.
The RAD51C gene encodes a protein that is essential for DNA repair. Variants of this gene that prevent the protein from working are known to increase the risk of breast and ovarian cancer and, in rare cases, if two harmful genetic changes are present, can lead to Fanconi anemia, a serious genetic disorder. Women who carry a defective RAD51C gene have a 15 to 30 percent lifetime risk of developing breast cancer and a 10 to 15 percent lifetime risk of developing ovarian cancer.
Although genetic testing is common among people with a strong family history of cancer, the health consequences of most RAD51C gene variants were previously unknown. This uncertainty about cancer risk often leaves patients and physicians unable to determine the appropriate medical care to provide.
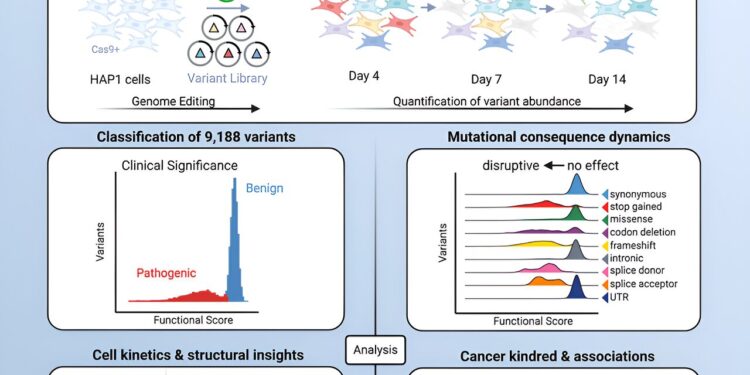
In this new study, researchers from the Wellcome Sanger Institute and their collaborators sought to understand the effect of 9,188 unique changes in the RAD51C gene by artificially altering the genetic code of human cells grown in a petri dish, in a process known as “saturation genome editing.” They identified 3,094 of these variants that can disrupt the gene’s function and increase cancer risk, with greater than 99.9% accuracy compared to clinical data.
Analysis of data from the UK Biobank and an ovarian cancer cohort of over 8,000 people confirmed the link between these harmful RAD51C gene variants and cancer diagnoses.
By mapping the protein structure, the team also identified crucial surface regions of RAD51C that are essential for its DNA repair function. These regions may interact with other yet-to-be-identified proteins or play a role in processes such as phosphorylation, providing valuable insights for drug development and potential new therapeutic targets.
The study also revealed the existence of “hypomorphic alleles,” a type of variant that reduces the function of the RAD51C gene without completely disabling it. These alleles appear to be more common than previously thought and may contribute significantly to breast and ovarian cancer risk.
Rebeca Olvera-León, first author of the study at the Wellcome Sanger Institute, said: “This research demonstrates that genetic risk for breast and ovarian cancer is not a simple yes or no scenario, but exists on a spectrum based on how genetic changes affect protein function. A more complete understanding of how genetic variants in the RAD51C gene contribute to cancer risk opens up new possibilities for more accurate risk prediction, prevention strategies and potentially targeted therapies.”
Dr Andrew Waters, co-senior author of the study at the Wellcome Sanger Institute, said: “This work demonstrates the power of analysing genetic variants on a large scale in their genomic context. Not only can we understand how cancer-related DNA changes affect patients, helping us make clinical decisions, but we can also explore how these variants impact gene function at a detailed molecular level. This provides important insights into how proteins work and how genes change over time.”
Dr David Adams, co-senior author of the study at the Wellcome Sanger Institute, said: “The strong link between harmful variants and cancer in large-scale studies suggests that this approach to classifying variants could be a valuable tool for personalised medicine and cancer prevention. We are keen to extend this technique to many more genes, with the aim of covering the entire human genome over the next decade with the Atlas of Variant Effects.”
Professor Clare Turnbull, clinical lead on the study, Professor of Translational Cancer Genetics at the Institute of Cancer Research, London and Consultant in Clinical Cancer Genetics at the Royal Marsden NHS Foundation Trust, said: “These new data will be very useful for diagnostic laboratories to better understand the changes in the RAD51C gene that we identify during clinical genetic testing in cancer patients and their family members.
“The data from the analysis will help us determine which genetic changes are harmful and which are harmless. This helps us make decisions about which patients might benefit from additional breast cancer screening and preventive ovarian surgery.”
More information:
Olvera-León, R. et al. High-resolution functional mapping of RAD51C by saturation genome editing, Cell (2024). DOI: 10.1016/j.cell.2024.08.039. www.cell.com/cell/fulltext/S0092-8674(24)00968-1
Cell
Provided by the Wellcome Trust Sanger Institute
Quote:Breast and ovarian cancer newly linked to thousands of genetic variants (2024, September 18) retrieved September 18, 2024, from
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without written permission. The content is provided for informational purposes only.