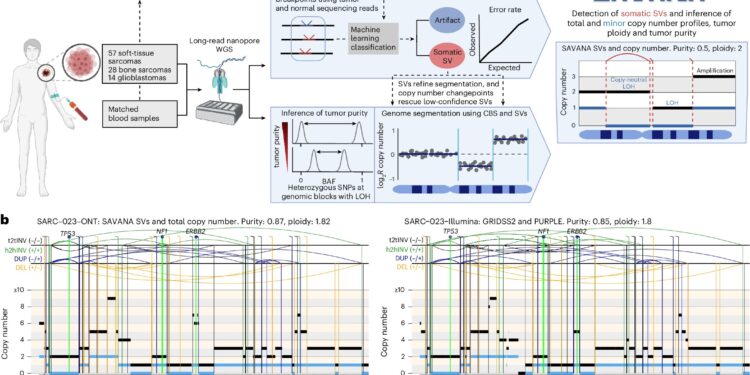
Savana overview. Credit: Nature methods (2025). DOI: 10.1038 / S41592-025-02708-0
Long -reading sequencing technologies analyze long DNA continuous expanses. These methods have the potential to improve researchers’ ability to detect complex genetic alterations in cancer genomes. However, the complex structure of cancer genomes means that standard analysis tools, including existing methods specifically developed to analyze long -reading wrestling sequencing data, are often shortened, leading to falsely positive results and unreliable interpretations of data.
These misleading results can compromise our understanding of the evolution of tumors, react to treatment and, ultimately, how patients are diagnosed and treated.
To meet this challenge, researchers have developed Savana, a new algorithm that they describe in the journal Nature methods.
Savana uses automatic learning to precisely identify structural variants – enlarged genomic alterations such as insertions, deletions, duplications or rearrangements – and the aberrations of the number of copies resulting in cancer genomes – using long -reading sequencing data.
It is important to have the right tool for work. For example, you can eat soup with a fork, but the result is not as effective as using a spoon. Savana, like a spoon, is suitable for the task and designed to effectively provide reliable results.
This algorithm was developed and tested on 99 samples of human tumors by researchers from the European Institute of Bioinformatics of EMBL’s EMBER (EMBL-EBI) and R&D Laboratory of Genomics England, in collaboration with the clinical partners of the University College London (UCL), of the Royal National OrthopeDic Hospital (RNOH), of the hospital Medicina João Lobo and the Boston hospital.
“Because other analysis tools are not developed to take into account the particularities of cancer genomic data, they often collect false positives that could lead to incorrect clinical and biological interpretations,” said Isidro Cortes-Ciriano, group leader at EMBL-EBI.
“Savana changes this. By dragging the algorithm directly on the long-reading sequencing data from cancer samples, we have created a new method that can make the difference between the real genomic alterations linked to cancer and the sequencing artefacts, thus allowing us to elucidate the mutational processes that underpin the cancer using a long-fight sequencing unprecedented resolution. “
Optimized for clinical use
“When we have developed Savana, our objective was clear: create a tool sufficiently sophisticated to characterize the genomes of complex cancer but sufficiently practical for clinical use,” said Hillary Elrick, former predoctoral comrade with EMBL-EBI and postdoctoral scholarship holder at the Francis Crick Institute.
“Consequently, Savana can precisely distinguish somatic structural variants, the aberrations of the number of copies, tumor purity and Ploidia – the key to understanding tumor biology and guiding clinical treatment decisions,” added Carolin Sauer, postdoctoral scholarship in EMBL -EBI.
Its rapid analysis and its correction of robust errors make Savana well suited to clinical use. The method has been recently applied to study osteosarcoma, rare and aggressive bone cancer that mainly affects young people, where it has helped researchers discover new genomic rearrangements, providing new information on how osteosarcoma is evolving and progressing.
The team also compared Savana’s results from long reading data with the illuminated sequencing of the same samples analyzed using a genome sequencing data analysis pipeline used to provide clinical reports.
The results were very coherent between technologies, demonstrating that Savana works on equal clinical standards while revealing additional cancer alterations.
“The ability to precisely detect structural variants is transformative for clinical diagnostics,” said Adrienne Flanagan, professor at UCL, a consultant histopathologist at RNOH.
“Savana could help us with confidence identifying the relevant genomic alterations for diagnosis and prognosis. In the end, this means that we would be better placed to offer personalized treatments for cancer patients.”
British investment in clinical genomics
The United Kingdom invests considerably in genomic sequencing technologies within the framework of the NHS Genomic Medicine Service. This initiative is the first in the world to offer a sequencing of the whole genome in the context of routine care. By integrating genomics into daily clinical practice, it aims to improve diagnostic precision and support personalized cancer treatments.
However, investments in clinical genomics will only reach their expected impact if the genomic data is interpreted with precision, which is based on specialized analytical tools. Genomics England explored the use of Savana as part of its work by examining the clinical potential of long reading sequencing technology to support a faster and faster diagnosis of cancer.
“The use of Savana will guarantee that clinicians receive precise and reliable genomic data, allowing them to integrate advanced genomic sequencing methods such as long reading sequencing in patients’ routine care,” said Greg Elgar, director of sequencing R&D at Genomics England.
Savana is also deployed in the context of national initiatives, such as the British Pediatric Pediatric Project of Stratified Medicine, co-directed by Cortes Ciriano. This project is focused on the development of more effective and less toxic treatments for childhood cancers using advanced sequencing technologies to better understand tumor biology and monitor the recurrence of diseases.
In addition, Savana is used in societal, ancestry, molecular and biological analyzes of inequalities (Sambai), a project aimed at treating the disparities in cancer in recent populations of African heritage.
More information:
Hillary Elrick et al, Savana: reliable analysis of somatic structural variants and aberrations of the number of copies using long reading sequencing, Nature methods (2025). DOI: 10.1038 / S41592-025-02708-0
Supplied by the European molecular biology laboratory
Quote: Automatic learning algorithm provides long bed sequencing to the clinic (2025, May 29) recovered on May 29, 2025 from
This document is subject to copyright. In addition to any fair program for private or research purposes, no part can be reproduced without written authorization. The content is provided only for information purposes.