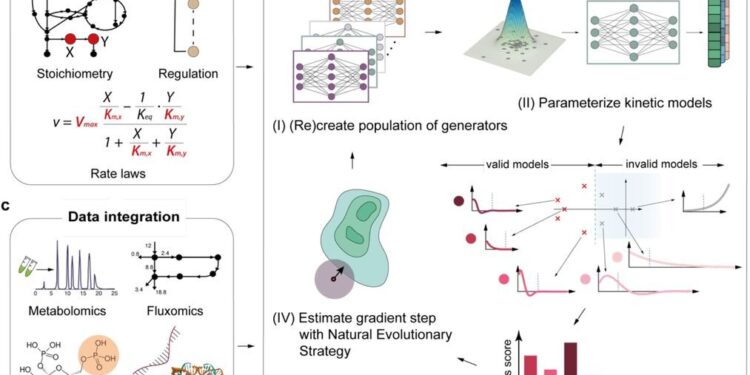
Overview and applications of the RENAISSANCE framework. Credit: Subham Choudhury et al.
Understanding how cells process nutrients and produce energy (metabolism) is essential in biology. Modern biology generates vast datasets on various cellular activities, but integrating and analyzing the vast amounts of data on cellular processes to determine metabolic states is a complex task.
Kinetic models help decode this complexity by providing mathematical representations of cellular metabolism. They act as detailed maps that describe how molecules interact and transform within a cell, illustrating how substances are converted into energy and other products over time. This helps scientists understand the biochemical processes that underlie cellular metabolism. Despite their potential, developing kinetic models is challenging because of the difficulty in determining the parameters that control cellular processes.
A team of researchers led by Ljubisa Miskovic and Vassily Hatzimanikatis from EPFL has just created RENAISSANCE, an artificial intelligence-based tool that simplifies the creation of kinetic models. RENAISSANCE combines different types of cellular data to accurately describe metabolic states, facilitating the understanding of how cells function. RENAISSANCE represents a major breakthrough in computational biology, opening new perspectives for research and innovation in the fields of health and biotechnology.
In their study published in Catalysis of natureThe researchers used RENAISSANCE to create kinetic models that accurately reflected the metabolic behavior of Escherichia coli. The tool successfully generated models that matched the metabolic behaviors observed experimentally, simulating how bacteria would adjust their metabolism over time in a bioreactor.
The kinetic models were also shown to be robust, maintaining their stability even when subjected to genetic and environmental perturbations. This indicates that the models can reliably predict the cellular response to different scenarios, thereby enhancing their practical utility in research and industrial applications.
“Despite advances in omics techniques, inadequate data coverage remains a persistent challenge,” says Miskovic. “For example, metabolomics and proteomics can only detect and quantify a limited number of metabolites and proteins. Modeling techniques that integrate and reconcile omics data from diverse sources can compensate for this limitation and improve systems understanding.”
“By combining omics data with other relevant information, such as extracellular media content, physicochemical data, and expert knowledge, RENAISSANCE allows us to accurately quantify unknown intracellular metabolic states, including metabolic fluxes and metabolite concentrations.”
RENAISSANCE’s ability to accurately model cellular metabolism has important implications, providing a powerful tool to study metabolic changes, whether induced by disease or not, and contributing to the development of new treatments and biotechnologies. Its ease of use and efficiency will enable a wider range of researchers from academia and industry to effectively use kinetic models and will foster collaboration.
More information:
Subham Choudhury et al, Generative machine learning produces kinetic models that accurately characterize intracellular metabolic states, Catalysis of nature (2024). DOI: 10.1038/s41929-024-01220-6
Provided by the Swiss Federal Institute of Technology in Lausanne
Quote:AI tool maps cellular metabolism with precision (August 30, 2024) retrieved August 30, 2024 from
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without written permission. The content is provided for informational purposes only.