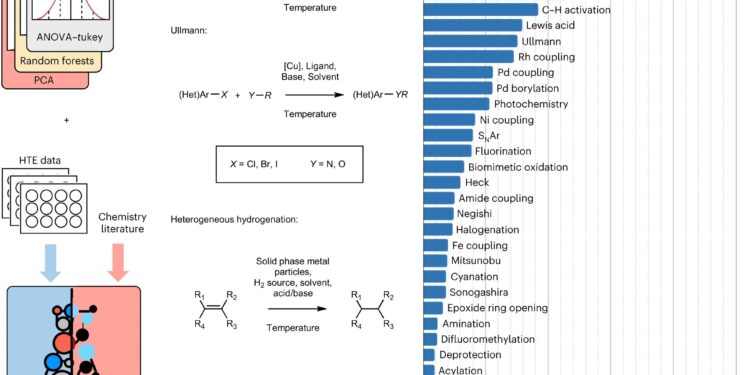
Overview of the HTE dataset and framework. A,An overview of HiTEA and its analysis is shown. Comparing the literature reactome with the HiTEA reactome will reveal support for our mechanistic conclusions (reactome agreement) or reveal areas of unusual chemical bias/phenomenon (reactome disagreement). bAbstract representations of the four classes of reactions analyzed by HiTEA in this publication are presented. vs,The distribution of the HTE dataset by reaction class is presented. Credit: Natural chemistry (2024). DOI: 10.1038/s41557-023-01393-w
Researchers have developed a platform that combines automated experiments with AI to predict how chemicals will react with each other, which could speed up the process of designing new drugs.
Predicting how molecules will react is vital to the discovery and manufacturing of new pharmaceuticals, but historically it has been a process of trial and error, and reactions often fail. To predict how molecules will react, chemists typically simulate electrons and atoms in simplified models, a computationally expensive and often inaccurate process.
Now, researchers at the University of Cambridge have developed a data-driven approach, inspired by genomics, in which automated experiments are combined with machine learning to understand chemical reactivity, significantly speeding up the process. They called their approach, which was validated on a dataset of more than 39,000 pharmaceutically relevant reactions, the chemical “reactome.”
Their results, reported in the journal Natural chemistryare the result of a collaboration between Cambridge and Pfizer.
“The reactome could change the way we think about organic chemistry,” said Dr Emma King-Smith of the Cavendish Laboratory in Cambridge, first author of the paper. “A deeper understanding of chemistry could allow us to make pharmaceuticals and many other useful products much more quickly. But more fundamentally, the understanding we hope to generate will benefit everyone who works with molecules.”
The reactome approach selects relevant correlations between reactants, reactants, and reaction performance from the data, and highlights gaps in the data itself. Data is generated from very fast or high-throughput automated experiments.
“High-throughput chemistry was a game-changer, but we believed there was a way to discover a deeper understanding of chemical reactions than can be observed from the first results of a high-throughput experiment,” he said. King-Smith said.
“Our approach reveals the hidden relationships between reaction components and outcomes,” said Dr. Alpha Lee, who led the research. “The dataset we trained the model on is enormous: it will help move the chemical discovery process from trial and error into the era of Big Data.”
In a related article, published in Natural communicationsThe team developed a machine learning approach that allows chemists to introduce precise transformations into predefined molecular regions, enabling faster drug design.
This approach allows chemists to modify complex molecules, such as a last-minute design change, without having to create them from scratch. Making a molecule in the laboratory is usually a multi-step process, like building a house. If chemists want to modify the nucleus of a molecule, the conventional method is to rebuild the molecule, like tearing down the house and rebuilding it from scratch. However, fundamental variations are important for drug design.
A class of reactions known as late-stage functionalization reactions attempt to directly introduce chemical transformations into the nucleus, thereby avoiding having to start from scratch. However, it is difficult to make functionalization selective and controlled at a late stage: many regions of the molecules can usually react, and it is difficult to predict the outcome.
“Late-stage functionalizations can produce unpredictable results and current modeling methods, including our own expert intuition, are not perfect,” King-Smith said. “A more predictive model would give us the ability to perform better screening.”
The researchers developed a machine learning model that predicted where a molecule would react and how the reaction site would vary under different reaction conditions. This allows chemists to find ways to precisely modify the nucleus of a molecule.
“We pre-trained the model on a large set of spectroscopic data, effectively teaching the general chemistry of the model, before refining it to predict these complex transformations,” King-Smith said. This approach allowed the team to overcome the limitation of low data: there are relatively few late-stage functionalization reactions reported in the scientific literature. The team experimentally validated the model on a diverse set of drug-like molecules and was able to accurately predict sites of reactivity under different conditions.
“The application of machine learning to chemistry is often limited by the problem of the small amount of data relative to the vastness of chemical space,” Lee said. “Our approach, which involves building models that learn from large data sets similar but not identical to the problem we are trying to solve, solves this fundamental challenge of small data and could unlock progress beyond functionalization at an advanced stage.”
More information:
Emma King-Smith et al, Probing the chemical “reactome” with high-throughput experimental data, Natural chemistry (2024). DOI: 10.1038/s41557-023-01393-w
Advanced Minisci predictive functionalization with transfer learning, Natural communications (2024). DOI: 10.1038/s41467-023-42145-1. www.nature.com/articles/s41467-023-42145-1
Provided by the University of Cambridge
Quote: Accelerating the manufacturing of new drugs using machine learning (January 15, 2024) retrieved January 15, 2024 from
This document is subject to copyright. Apart from fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.