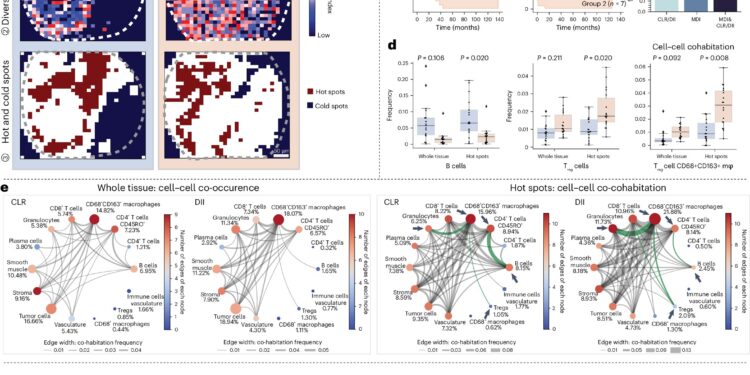
The multiomic and ecological analysis improve the prognostic capacities of the CRC. Credit: Nature genetics (2025). DOI: 10.1038 / S41588-025-02119-Z
To understand what leads to the progression of the disease in the tissues, scientists need more than a simple instantaneous cells in isolation – they need to see where the cells are, how they interact and how this spatial organization moves between the pathological states. A calculation method called MESA (Multioomic and ecological spatial analysis), detailed in a study published in Nature geneticsHelp researchers to study sick tissues more significantly.
The work details the results of a collaboration between MIT researchers, the University of Stanford, Weill Cornell Medicine, the Ragon Institute of MGH, MIT and Harvard, and the Broad Institute of Mit and Harvard, and was led by the Stanford team.
Mesa brings a lens inspired by ecology to tissue analysis. It offers a pipeline to interpret spatial ommal data – the product of advanced technology that captures molecular information as well as the location of cells in tissue samples. These data provide a high resolution card of the “districts” of fabrics and Mesa helps to give meaning to the structure of this card.
“By integrating traditionally distinct approaches to disciplines, Mesa allows researchers to better appreciate how the tissues are organized locally and how this organization changes into different contexts of diseases, propel new diagnostics and the director of the Institute for Medical Engineering and Sciences (IMES), the professor of the JW KIECCH Koch Institute for Integutive Cancer Research at MIT, as well as a member of the Institute Broad Institute and member of the Ragon Institute.
“In ecology, people study biodiversity between regions – how distributed animal species are and interact,” explains Bokai Zhu, Postdoc and author of the study. “We realized that we could apply these same ideas to tissue cells. Instead of rabbits and snakes, we analyze T cells and B cells.”
Presentation of Mesa. Credit: Nature genetics (2025). DOI: 10.1038 / S41588-025-02119-Z
By treating the types of cells like ecological species, Mesa quantifies “biodiversity” in tissues and follows the way in which this diversity changes its disease. For example, in liver cancer samples, the method has revealed areas where tumor cells co-coïraged with macrophages, which suggests that these regions can generate unique disease results.
“Our method reads tissues like ecosystems, discovering cellular” hot spots “that mark the first signs of disease or response to treatment,” adds Zhu. “This opens up new possibilities for diagnostic for precision and therapy design.”
Mesa also offers another major advantage: it can enrich tissue data in calculation without the need for more experiences. Using single -cell data sets accessible to the public, the tool transfers additional information, such as gene expression profiles – on existing tissue samples. This approach deepens the understanding of the functioning of space domains, in particular when comparing healthy and sick tissues.
In tests on several data sets and tissue types, Mesa has discovered spatial structures and key cell populations that were previously neglected. It incorporates different types of OME data, such as transcriptomic and proteomics, and builds a multilayer view of tissue architecture.
Currently available as a Python package, Mesa is designed for university and translational research. Although spatial ommics is still too much with a high intensity of resources for clinical use in routine hospital, technology is gaining ground among pharmaceutical companies, especially for drug tests where understanding of tissue responses is essential.
“It’s just the start,” said Zhu. “Mesa opens the door to the use of ecological theory to disentangle the spatial complexity of the disease – and finally, to better predict and treat it.”
More information:
Daisy Yi Ding et al, quantitative characterization of tissue states using a multiomic and ecological spatial analysis, Nature genetics (2025). DOI: 10.1038 / S41588-025-02119-Z
Supplied by the Massachusetts Institute of Technology
This story is republished with the kind authorization of Mit News (Web.mit.edu/newsoffice/), a popular site that covers news of research, innovation and MIT teaching.
Quote: A new calculation framework illuminates the hidden ecology of sick tissues (2025, April 28) recovered on April 28, 2025 from
This document is subject to copyright. In addition to any fair program for private or research purposes, no part can be reproduced without written authorization. The content is provided only for information purposes.