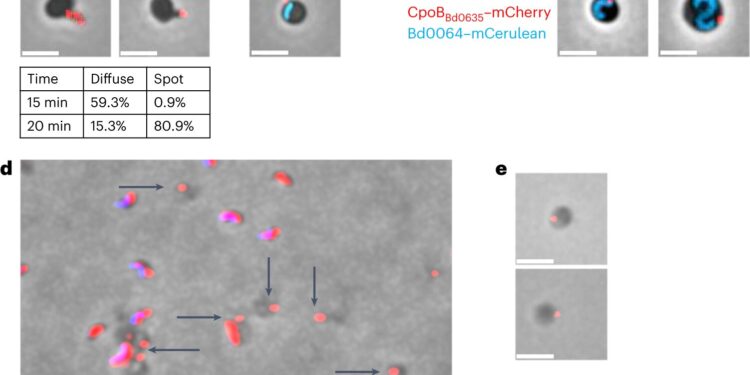
B. bacteriovorus deposits a robust vesicle “invasion organelle” during invasion. Credit: Natural microbiology (2024). DOI: 10.1038/s41564-023-01552-2
A decades-old mystery about how naturally occurring antimicrobial predatory bacteria are able to recognize and kill other bacteria may have been solved, new research suggests.
In a study published in Natural microbiologyResearchers from the University of Birmingham and the University of Nottingham have discovered how naturally occurring antimicrobial predatory bacteria, called Bdellovibrio bacterivores, produce fiber-like proteins on their surface to trap their prey.
This discovery could allow scientists to use these predators to target and kill problematic bacteria that cause health, food spoilage and environmental problems.
Andrew Lovering, professor of structural biology at the University of Birmingham, said: “Since the 1960s, the bacterivore Bdellovibrio has been known to hunt and kill other bacteria by entering target cells and eating them from the inside out. before bursting later. The scientists wondered, “How can these cells attach tightly when we know how varied their bacterial targets are?” »
Professor Lovering and Professor Liz Sockett, from the School of Life Sciences at the University of Nottingham, have been collaborating in this area for almost 15 years. The breakthrough came when undergraduate Sam Greenwood and Asmaa Al-Bayati, a Ph.D. student in the Sockett lab, discovered that Bdellovibrio predators deposit a solid vesicle (a “pinched” part of the predator’s cell envelope) when they invade their prey.
Professor Liz Sockett explained: “The blister creates a sort of airlock or keyhole allowing Bdellovibrio to enter the prey’s cell. We were then able to isolate this vesicle from the dead prey, which is a first in this field. The vesicle was analyzed to reveal tools used during the previous predator/prey contact event. We thought of it a bit like a locksmith leaving the pick, or key, as evidence, in the keyhole.
“By examining the contents of the vesicles, we discovered that because Bdellovibrio does not know which bacteria it will encounter, it deploys a range of similar prey recognition molecules on its surface, creating many different ‘keys’ for it.” unlock “many different molecules. prey types.”
The researchers then carried out an individual analysis of the molecules, demonstrating that they form long fibers, approximately 10 times longer than common globular proteins. This allows them to operate from a distance and “feel” nearby prey.
In total, the laboratories counted 21 different fibers. Researchers Dr Simon Caulton, Dr Carey Lambert and Dr Jess Tyson have worked on how they work at the cellular and molecular levels. They were supported by genetic engineering of fibers by Paul Radford and Rob Till.
The team then began attempting to connect a particular fiber to a particular molecule on the surface of prey. Discovering which fiber corresponds to which prey could enable a technical approach allowing tailor-made predators to target different types of bacteria.
Professor Lovering continued: “Because the predator strain we are studying comes from the ground, it has a broad killing spectrum, making the identification of these fiber and prey pairs very difficult. However, on the fifth attempt to find the partners, we discovered a chemical. signature on the outside of the prey bacteria that fitted tightly to the end of the fiber. This is the first time that a characteristic of Bdellovibrio has been adapted for prey selection.
Scientists in this field will now be able to use these findings to ask which set of fibers are used by the different predators they study and potentially assign them to specific prey. Better understanding these predatory bacteria could allow their use as antibiotics, to kill bacteria that spoil food or those that are harmful to the environment.
Professor Lovering concluded: “We know that these bacteria can be useful, and by fully understanding how they work and find their prey, it opens up a world of new discoveries and possibilities. »
More information:
Caulton, SG et al, Bdellovibrio bacteriovorus uses chimeric fiber proteins to recognize and invade a wide range of bacterial hosts, Natural microbiology (2024). DOI: 10.1038/s41564-023-01552-2 www.nature.com/articles/s41564-023-01552-2
Provided by the University of Birmingham
Quote: Scientists solve the mystery of how predatory bacteria recognize their prey (January 4, 2024) retrieved January 4, 2024 from
This document is subject to copyright. Apart from fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.