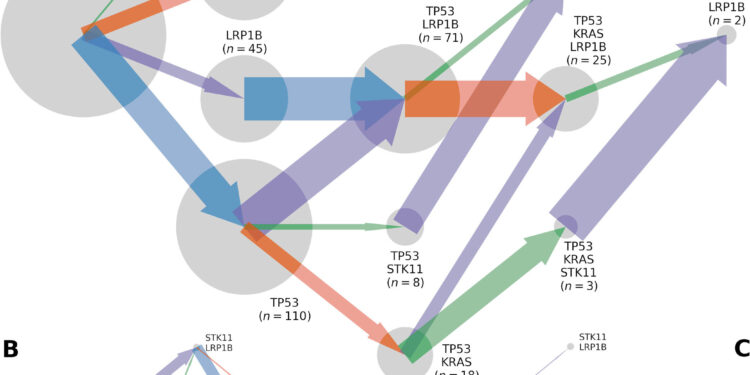
Trajectories of somatic evolution by mutation of TP53, KRAS, LRP1B and STK11, inferred from a total of 565 lung adenocarcinoma tumors sequenced in the whole exome. Somatic genotypes (gray circles; areas are proportional to the n observed for the somatic genotype) evolve according to (A) fluxes, (B) mutation rates, and (C) selection coefficients proportional to the width of the arrows pointing to a somatic genotype. to another, colored by the gene in which the mutation occurs (TP53, blue; KRAS, orange; STK11, green; LRP1B: purple). Credit: Mathematical Biosciences (2023). DOI: 10.1016/j.mbs.2023.109091
Cancer occurs when multiple mutations lead to incessant and inappropriate cell growth. But these mutations do not act in isolation. Instead, mutations can influence each other in ways that affect cancer progression. Researchers have long struggled to understand these interactions, often making assumptions that oversimplify the complex reality.
A new method from the Yale School of Public Health (YSPH) offers a way to analyze how mutations interact with each other to alter the development of tumors. Innovation should make it easier to develop targeted therapies that anticipate the evolution of a cancer, then trap it and eradicate it.
“We can now characterize where cancer is on its genetic trajectory in a given patient,” said lead author Jeffrey P. Townsend, Elihu Professor of Biostatistics at the Yale School of Public Health and professor of ecology and biology. evolving at Yale. “This information can be very helpful in determining appropriate treatments, especially as we get more and more options for precision treatment of tumors.”
The results are published in Mathematical biosciences.
How much blame should be attributed to each mutation?
To become cancerous, cells mutate and develop characteristics called cancer hallmarks. These characteristics include the ability to generate growth signals or ignore signals to stop growth, to metastasize, to generate new blood vessels to serve the tumor(s), to dodge immune cells that can detect and kill a aberrant cell, etc. Cancer cells can mutate in various ways to acquire these characteristics. Once a cancerous cell, it continues to evolve.
This continued adaptation to their environment makes cancer difficult to treat in a targeted manner. A targeted drug creates evolutionary pressure: surviving cells quickly come to predominate in the tumor, ultimately rendering the drug ineffective. A way to predict which mutations are likely to occur next could help clinicians find ways to prevent resistance.
A few years ago, Townsend and his colleagues devised a way to estimate the importance of each mutation in a cancer by examining the frequency of each individual mutation in a large number of tumors as well as the underlying rate at which this mutation appears.
“This was a major breakthrough, because before this, everyone was calling mutations ‘carcinogenic’ or not, but not quantifying the contribution of each mutation,” said Townsend, who is also affiliated with the Computational Biology Program and in Bioinformatics from Yale.
Cancers have multiple mutations. The next step was to characterize not only the average effect of each mutation, but also how each interacts with the next one that occurs.
The term for these interactions is epistasis: the way one mutation affects the extent to which another mutation allows a cancer to grow and survive. Unraveling epistasis is complex, especially when considering the relationships between three or more mutations.
For the current project, Townsend began by deriving a mathematical approach to estimate epistasis for pairs of point mutations. He then teamed up with Jorge Alfaro-Murillo, a research associate in biostatistics at YSPH, who is the first author of the study.
Alfaro-Murillo developed a mathematical approach that, with enough data, will provide estimates of epistatic interactions between three, four, or even more mutations.
The order of mutations is important
Researchers have long noticed that some mutations always seem to coexist in a given cancer, while others seem to be mutually exclusive. For this reason, many previous studies have assumed that certain mutations work together or oppose each other.
But this is not necessarily the case, because not all co-occurrences are true biological interactions. For example, some might arise because a certain exposure, such as tobacco smoke, tends to result in characteristic mutations, each appearing independently due to the smoke itself.
“There are tons of approaches to looking at mutual exclusivity and co-occurrence and trying to determine how often they occur in sets of tumors. But mutual exclusivity and co-occurrence is just not the best way to determine mutual exclusivity and co-occurrence. response,” Townsend said. “Our method gives a better answer to the question of which genes interact.”
In addition to accounting for underlying mutation rates, “it does so in part by taking into account the order in which mutations occur,” Alfaro-Murillo explained.
For example, let’s say that the job of gene A is to cause a dangerously mutated cell to self-destruct, while the job of gene B is to cause a cell to multiply.
If a cell first develops a mutation in gene B, then a normal gene A will ensure that the cell dies before it divides uncontrollably. But if gene A mutates first, followed by gene B, the cell can survive and begin to multiply. Order matters.
“If the first A mutation occurs, then the B mutation, which could be more important than the B mutation preceding A,” Alfaro-Murillo said. “This is a major difference from simply looking at mutual exclusivity.”
Translating findings into cancer care
In an important limitation, the authors considered only tumors that had not been exposed to treatments. They then plan to look at tumors responding to treatment, as well as look beyond point mutations and consider mutations that cause larger changes, such as copy number changes in the genome or chromosomal alterations. Copy number alterations, also known as CNAs, are somatic changes in chromosomal structure that result in gain or loss of copies of sections of DNA and are prevalent in many types of cancer.
These analytical methods should help make cancer trials and, ultimately, treatments involving multiple anticancer drugs more efficient.
“If your tumor has a certain composition of mutations and you know that a treatment increases the likelihood that you will get certain mutations, then if there are drugs targeted for those mutations, perhaps you could apply them immediately,” he said. Alfaro-Murillo said. said. “If you can see what is most likely to happen next, then you can prepare for it.”
More information:
Jorge A. Alfaro-Murillo et al, Pairwise and higher-order epistatic effects among somatic cancer mutations through oncogenesis, Mathematical Biosciences (2023). DOI: 10.1016/j.mbs.2023.109091
Provided by Yale University
Quote: Analyzing how cancer mutations interact can improve targeted therapies (January 2, 2024) retrieved January 2, 2024 from
This document is subject to copyright. Except for fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.