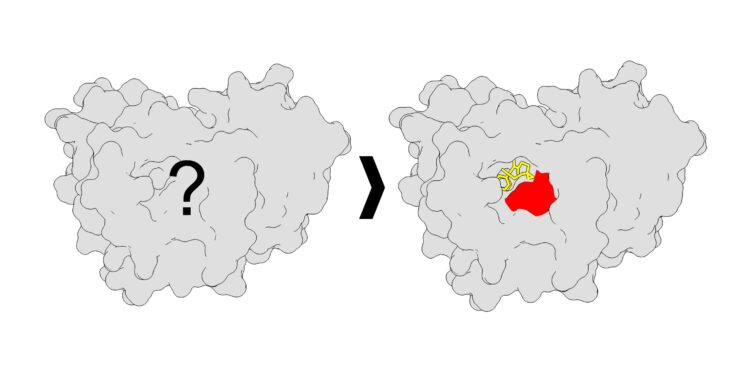
A protein with unknown binding sites (left), versus a protein with high-resolution binding site mapping (right). Credit: Scripps Research
Identifying new ways to target proteins involved in human diseases is a priority for many researchers around the world. However, discovering how to change the function of these proteins can be difficult, especially in living cells. Now, scientists at Scripps Research have developed a new method to examine how proteins interact with small drug-like molecules in human cells, revealing crucial information about how to potentially target them therapeutically.
The strategy, published in Nature Chemical Biology on January 2, 2024, uses a combination of chemical and analytical techniques to reveal the specific places where proteins and small molecules bind together. Ultimately, this method could lead to the development of more targeted and effective therapies.
“Our new technology could be used to find new druggable sites on proteins for any human disease, from cancer to Alzheimer’s disease,” says Christopher Parker, Ph.D., associate professor in the Department of Chemistry and lead author of the study. “We have no restrictions on how this could be used. Our work has the potential to usher in a whole new way of drug discovery.”
The Parker lab aims to uncover how proteins work in each human cell type in order to develop effective treatments for a wide range of human diseases. In this study, Parker and his team built on their initial work in the laboratory of Professor Benjamin Cravatt at Scripps Research to create a new method for examining how proteins interact with small molecules in living cells.
They developed an analytical strategy to better understand how these proteins interact with small molecules at a much higher resolution than ever before. To do this, they used chemical probes called photoaffinity probes, which are molecules that can be activated by light to allow the probes to capture a bound protein.
Credit: Nature Chemical Biology (2024). DOI: 10.1038/s41589-023-01514-z
By gathering data on protein interactions with photoaffinity probes, the Parker team identified locations on proteins where small molecules could connect and bind. Essentially, the team discovered more than a thousand new locks (binding sites on proteins) and corresponding keys (small molecules), the vast majority of which were new small molecule binding loci that had not been previously discovered. previously reported. Additionally, they discovered new features of the binding sites, such as new shapes.
“Identifying these specific binding sites will help scientists design new molecules that fit even better into these pockets, potentially leading to more effective therapies,” says Jacob M. Wozniak, co-first author and former researcher. postdoctoral fellow in the Parker laboratory. The paper’s other co-first author was Weichao Li, Ph.D., a research associate also in the Parker lab.
Using the wealth of data from this study and collaborating with co-author Stefano Forli, Ph.D., associate professor in the Department of Integrative Structural and Computational Biology, the authors then modeled how certain molecules might bind to these proteins. This library of information could be used to design treatments that interact with proteins in more targeted ways.
“Our new process reveals additional opportunities for therapeutic intervention and discovery in human cells,” says Parker. “Next, we plan to use this technology to target proteins relevant to autoimmune diseases and cancer.”
More information:
Jacob M. Wozniak et al, Improved mapping of small molecule binding sites in cells, Nature Chemical Biology (2024). DOI: 10.1038/s41589-023-01514-z
Provided by the Scripps Research Institute
Quote: New mapping method sheds light on drug sites on proteins (January 2, 2024) retrieved January 2, 2024 from
This document is subject to copyright. Except for fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.